Grantee Research Project Results
2016 Progress Report: Vanderbilt - Pittsburgh Resource for Organotypic Models for Predictive Toxicology
EPA Grant Number: R835736Center: Mickey Leland National Urban Air Toxics Research Center (NUATRC)
Center Director: Beskid, Craig
Title: Vanderbilt - Pittsburgh Resource for Organotypic Models for Predictive Toxicology
Investigators: Hutson, Michael Shane , Osteen, Kevin G. , McCawley, Lisa J. , Bruner-Tran, Kaylon L. , Taylor, D. Lansing , Davidson, Jeffrey M. , Gough, Albert , Cliffel, David , Aronoff, David , Markov, Dmitry , Wikswo, John , McLean, John , Vernetti, Lawrence , Shotwell, Matt , Tuan, Rocky
Current Investigators: Tuan, Rocky , Alexander, Peter
Institution: Vanderbilt University , University of Pittsburgh
Current Institution: University of Pittsburgh , Vanderbilt University
EPA Project Officer: Callan, Richard
Project Period: December 1, 2014 through November 30, 2018 (Extended to November 30, 2019)
Project Period Covered by this Report: December 1, 2015 through November 30,2016
Project Amount: $6,000,000
RFA: Organotypic Culture Models for Predictive Toxicology Center (2013) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
This center will advance predictive toxicology by developing four organotypic culture models (OCMs): liver, mammary gland, limb/joint development, and fetal membrane. These 3D cultures of heterotypic cells will better approximate the in vivo cellular microenvironment and are amenable and affordable for medium to high throughput screening. Screening of these OCMs against 30-50 chemicals from the ToxCast inventory will demonstrate whether OCM toxicity is predictable from ToxCast in vitro assays and whether OCM toxicity is predictive of previous animal studies and/or human epidemiology.
Progress Summary:
In Year 2, the VPROMPT Center has:
- Successfully developed first generation (Gen1.0, including six parallel, 4-chambers each) mammary gland bioreactors for Project 1. Upcoming expansion of the Gen1.0 mammary bioreactor into a second generation (Gen2.0) mammary OCM with active perfusion using Project 5 hardware already is underway. The mammary gland monocultures have been fully optimized and medium throughput validation has been started.
- Established differentiation constructs for chondrogenesis (OCM1) and for hypertrophy (OCM2) as part of limb development in Project 2. Characterization of chondrogenesis and hypertrophy, including the determination of detection limits and baseline values as well as assessment of variability among cultures for each assay using these small volumes is complete. We have concluded testing of three known teratogens and two putative teratogens (BPA, Mifepristone) using OCM1 and OCM2. Project 2 bioreactors currently are undergoing integration with hardware for microfluidic perfusion purposes.
- Project 3 has established dosage and exposure times for LPS and TCDD and validated inflammatory biomarkers in transwell biopsies for fetal membrane device development in Project 3. The fetal membranes team is finalizing establishment of the IFMOC 3.0 by validating OCM responses to selected toxicants before repeating LPS studies in the IFMOC.
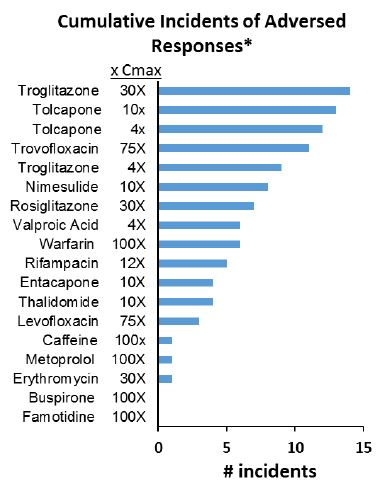
- The liver project (Project 4) was able to demonstrate secretome screening via IM-MS of their liver OCM devices exposed to tolcapone, entacapone, and vehicle controls. In addition, four studies of 18 day duration for 15 test agents for the EPA have been completed. Results showed that out of a possible 29 toxicity measurements collected over an 18-day treatment period, troglitazone produced the highest cumulative total of adverse events compared to other compounds. Clinical hepatotoxins troglitazone, tolcapone, trovafloxacin, and nimesulide produced more adverse incidents than compounds linked to iDILI (idiosyncratic drug-induced liver injury), such as valproic acid and warfarin, or when compared to nonhepatotoxins like caffeine or buspirone. For the liver OCM, an adverse event was defined as a 50% change from control for any of the following measurements: LDH (N = 18), albumin (3), urea (3), cytochrome C biosensor (2), bile efflux (1), TNF-α (1), and testosterone clearance (1).
Further, we have successfully adapted the University of Pittsburgh Microphysiology Database (UPMDb) to manage, consolidate, visualize, QC, and organize OCM data for transfer to the EPA and we already have compiled pre-clinical, clinical, and epidemiology findings on VPROMPT chemicals into UPMDb.
Finally, our systems engineering & analysis teams (Project 5) continue to deliver standalone pumps, Perfusion Controller (IOM-PC, v1.0), and MicroClinical Analyzer (μCA, v1.0) hardware integration modules, as well as different limb segmentation (OCM3) prototypes as requested by organ module developers. The MicroClinical Analyzer electrodes, fluidics, and housing have been redesigned to reduce sensor volume to 6 μl, while increasing the number of testable analytes from five to eight. A 26-position rotary planar valve (RPV) was developed for our Microformulator v2.0. Finally, our flexible and powerful OCM hardware control software now enables full automation of integrated modules and we are well underway integrating wireless capabilities to support these modules.
Project 1 (Mammary Gland): During Year 2, the team has developed a monoculture mammosphere OCM in a six device, parallel testing system (Gen1.0) for medium throughput screening. They currently are finalizing integration of perfusion hardware (Gen2.0) and evaluation of the tri-culture system. Detailed protocols have been developed for toxicant interactions with PDMS, including saturation tests and on/off rates where interaction was noted. This investigation is especially valuable as several microfluidic organotypic cell culture systems are fabricated using this material. Further, Project 2 has produced stably-transfected cell lines with fluorescent, lentiviral biosensors in order to allow for medium-throughput microscopy screening of toxicant effects. Finally, 3D culture methods including measurable biomarker readouts for protease activity have been established. Because at life stages of active tissue growth and remodeling the mammary gland is particularly sensitive to toxicant exposures, this OCM developed for screening and evaluation of ToxCast chemicals will be especially valuable for environmental research.
Project 2 (Limb Development): In Year 2, Project 2 tasked itself with adapting established 20-μL cultures comprising 400,000 cells culture to a medium throughput format. This involved: (a) reducing the cell number, (b) developing non-invasive analytical methods for analysis, and (c) changing the culture vessel to accommodate microfluidics.
- Reducing the scale of the culture: Conditions required for establishing 2-μL, 40,000-cell micromass cultures were determined including texturization and collagen I coating of the polystyrene growth surface and shortening the adhesion phase to 30 minutes. Thereafter, Project 2 verified that 40,000-cell cultures undergo chondrogenesis (OCM1) and hypertrophy (OCM2) in a manner analogous to the process observed in vivo and in the 400,000-cell cultures reported in Year 1. Ultimately, the texturized surface will be replaced by porous, inert membranes such as polycarbonate (see below). With the smaller cultures, critical time points for analysis in the time frame of both chondrogenic and hypertrophic cultures can be reduced.
- Developing non-invasive analytical methods: Project 2 has tested several promoter-reporter constructs for signal transduction and differentiation pathways indicating cell stress and death as well as stages of skeletal development: chondrogenesis, hypertrophy, and joint segmentation. This included optimization of plasmid transfection and lentiviral transduction conditions, demonstration of promoter function in adult human MSCs, and demonstration of normal differentiation following lentiviral transduction. These experiments have provided preliminary evidence that some toxicants, specifically mifepristone, may activate specific signaling pathways, i.e., canonical WNT signaling mediated via β-catenin.
- Changing the culture vessel to accommodate microfluidics: Project 2 is collaborating with P5 to develop a microfluidically-enabled bioreactor for OCM1 and 2. These bioreactors will be derived from an existing brain-on-a-chip model developed by members of Project 5. In this model, the cells can be grown on a porous membrane of polycarbonate. As proof of concept, Project 2 has prepared chondrogenic cultures on porous polycarbonate transwell inserts and observed excellent cartilage matrix elaboration. Furthermore, Project 2 has determined that the Kiyatec Cube™ system will serve to optimize culture surface topography and flow conditions to support OCM development. In anticipation of eventual connection of the limb models with the liver OCM in Project 4, Project 2 has determined that the liver medium formulation (plus OCM specific differentiation supplements) does not change OCM1 and OCM2 development.
Project 2 continued to work in close collaboration with Project 5 to develop a mechanostimulatory bioreactor for modeling joint segmentation (OCM3). Project 5 has delivered several versions of a concentric ring-based bioreactor (version 1). Although surface conditions, seeding procedures, and differentiation conditions were tested and optimized by Project 2, it was determined that the requirements of the bioreactor dimensions and manufacture (as determined by the biology of the seeded cells) made fabrication of this device at a suitable scale very difficult. A second, simpler and smaller bioreactor is under development (version 2) involving a cartilage rod that is immobilized within an anchoring block. Flexion is produced using an actuator that moves the free end of the cartilage rod submerged in medium. This model is more easily scalable to medium throughput testing.
Altogether, the results Project 2 has collected this past year will be useful in moving VPROMPT closer toward limb development organ-chip models in suitable bioreactors capable of elucidating the details of adverse outcome pathways from molecular toxicant initiating events to tissue-level responses in order to protect human health.
Project 3 (Fetal Membranes): The Fetal Membranes project team has successfully tested experimental concentrations of lipopolysaccharides (LPS, 10 ng/ml) in combined use with TCDD (10 nM) for an exposure of 4 hours (LPS exposure) and 24 hours (TCDD exposure). Inflammatory biomarkers to be used are IL-1 beta, TNF-alpha, IL-6, and IL-8, which all have been validated using fetal membrane punch biopsies via cytometric bead array panels, PCR, and ELISA assays. When amniotic cells were primed with TCDD, the team found that MMP7 levels decreased and MMP8 levels increased. Similarly, proteinases for collagen type IV (MMP2 and MMP9) are dysregulated by TCDD exposure. Most significantly, observations using the transwell-based fetal membrane model revealed that the sidedness of exposure was not a critical determinant of response in the context of toxicant screening and hence Project 2 chose to use punch membrane biopsies as an appropriate model for the initial screening studies.
At the same time, Project 2 has been developing the IFMOC 3.0 multilayer microfluidic model. They were able to demonstrate tight junction formation of human amniotic epithelial cells (hAMEpi) inside the device and showed that FITC-dextran diffusion across the membrane could thus be prevented. Co-cultures with hAMEpi cells already have been successfully maintained for 7 days inside a two-chamber IFMOC device.
Because fetal membrane inflammation is a leading cause of preterm birth, it is important to develop a model that can recapitulate various inflammatory responses. Using a mouse model, we have previously shown that exposure to environmental toxicants (TCDD) can cause an even more exaggerated inflammatory response to such microbial infections. Hence, our work to also demonstrate these fetal membrane behaviors in the transwell and IFMOC devices is very important for OCM validation and later ToxCast chemical evaluation.
Project 4 (Liver): The liver OCM was developed and previously characterized in the Nortis commercial microfluidic device (Seattle, WA) at the Drug Discovery Institute laboratory of D. Lansing Taylor, University of Pittsburgh. The liver OCM consists of four human liver cell types (primary human hepatocytes, a Kupffer-like immune cell, stellate, and endothelial cells) that are constructed as a 3D, microfludic tissuelike liver structure modeling the liver acinar organization. A subset of the human hepatocytes was transduced with a lentiviral construct to encode the hepatocyte genome with an expressible fluorescent cytochrome C protein in the mitochondria to monitor activation of apoptosis. Construction and use of the model and fluorescent protein transductions have been previously published.
In 2016, we used the Liver OCM to compare the influx and efflux media for secretome substrates and metabolites of over 1600 compounds from biomolecules including but not limited to fatty acids; amino acids and dipeptides; carbohydrates; small organic acids; catecholamines; oils, lipids; pyrroles; and polyamines. MetaboAnalyst 3.0 analysis of the m/z and MS/MS fragmentation products obtained from IM-MS/MS secretome measurements found four unique pathways affected by tolcapone including histidine metabolism, propanoate metabolism, protein biosynthesis, and valine, leucine and isoleucine degradation. The unique pathways affected by entacapone included aspartate metabolism, citric acid cycle, fructose and mannose degradation, galactose metabolism, mitochondrial electron transport chain, and phenylalanine and tyrosine metabolism. Additional analysis of results is planned to better understand the findings.
Seventeen compounds were tested at 1 or 2 concentrations for 18 days of treatment in the liver OCM. The cumulative incidents of adverse responses (an adverse response is defined as a 50% change from control for any of the following measurements: 18 LDH, 3 Albumin, 3 Urea, 2 Cytochrome C biosensor, and 1 each for Bile Efflux, TNF-α and Testosterone clearance measurements) for the compounds as shown in the figure. Out of a possible 29 toxicity measurements collected over the 18 day treatment period, the clinical hepatotoxins troglitazone, tolcapone, trovafloxacin, and Nimesulide produced more adverse incidents then iDILI compounds such as valproic acid and warfarin or when compared to non-hepatotoxins caffeine or buspirone.
The third milestone, to incorporate the liver OCM into an integrated platform being developed at Vanderbilt University, will be completed in 2017.
Summary Implications to EPA mission. The data generated from the secretome and the compound screening are expected to be used in conjunction with computational assessment at the University of Pittsburgh and EPA to develop tools that would better assess in vivo human risk from exposure to a chemical or metabolites. The liver OCM is included in this project for two reasons: a) to provide a platform that bioactivates test agents to study the effect of metabolites on the developmental organs in the project; and b) to provide time dependent changes in liver functions, clearance, initiation of apoptosis, or activation of liver immune cells as a response to compounds with known mechanisms of toxicity. These data are expected to produce experimental evidence to better define adverse outcome pathways associated with compound treatment.
Project 5 (Systems Engineering & Analysis): Project 5 teams are responsible for development of microfluidic perfusion hardware that will allow multiple organotypic culture models to be integrated with each other and electrochemical sensor systems (MicroClinical Analyzer) that will enable evaluation of OCM health prior to and after toxicant exposure. Glutamate sensors have been developed for the MicroClinical Analyzer. In addition, cholesterol and ammonium sensors are being finalized. The sensor volume has been reduced to 6 μl. We also have refined our processes to produce circular-segment 5-port and 24-port rotary planar valves (RPVs) and rotary planar peristaltic micropumps (RPPMs) that span a range of flow rates from 0.5 to 800 μL/min. At the same time, the team has made excellent progress toward full software automation of OCMs and integration of wireless capabilities. Both will greatly reduce the time required for hands-on OCM manipulations and increase screening throughput. Finally, the hardware team routinely is producing prototypes for limb development segmentation devices (OCM3, Project 2).
The ion mobility-mass spectrometry team of Project 5 has established baseline metabolic readouts for the liver OCM and is ready to establish baselines for Projects 1 and 3 as these devices reach a finalized development stage.
Finally, the prediction modeling team has recommended a uniform protocol for determining AC50s and maximal responses based on nonlinear least squares regression methods. Potential methodologies for clustering metrics and for constructing meaningful prediction models have been discussed. As training data from OCM chemical exposure experiments become available, the team anticipates an increase in their efforts under this milestone.
Future Activities:
Project 1 will continue collaboration with Project 5 toward upgrading the platform integrated MG OCM to include smaller footprint valves and pumps necessary for sampling and coupling the liver OCM upstream of the MG OCM. Further, we will be using measures for on/off rates and saturation values and generate a mathematical description of toxicant/bioreactor interaction. We also plan to screen toxicants, validate the MG OCM reactor system for medium throughput for stage specific and acute toxicity assessment using optical and electrochemical sensing, and move towards High Content profile analysis beginning with proteinase activity analysis of mammosphere formation using an identified toxicant. Finally, we will develop strategies for liver OCM-MG OCM coupling.
Project 2 plans to complete its QAPP and gain IACUC approval for a protocol for dosing of rat dams with teratogens. Using OCM1 and OCM2 (for which chondrogenesis and hypertrophy have been characterized by RT-PCR and histology) we will:
a) Complete assessment of signal transduction promoter-reporter constructs and differentiation stage-specific promoter-reporter constructs. Completion will include a correlation between promoter activity (the surrogate) and actual target gene activity under control conditions and a means to quantify promoter activity.
b) Test the utility of matrix anabolic processing and catabolic degradation products from the medium using ELISAs.
Further, we will continue to work on OCM3 (joint segmentation). We will test/develop a system for generating cartilage rods of appropriate length and diameter to accommodate a model that can be flexed appropriately. Once the model is developed, we will begin characterization of tissue development by RT-PCR, histology and promoter-reporter activity. In addition, we will develop microfluidically-enabled bioreactors for OCM1 and OCM2:
a) First, we will test OCM1 and OCM2 in a proof-of-concept platform based on the Kiyatec Cube using solid and porous PCL membranes and syringe pumps. The important part in this model will be establishing that the 2 μl culture can undergo normal chondrogenic and hypertrophic differentiation (as determined by RT-PCR and histology) on the membranes under flow. Membrane type and topography as well as flow rates will be communicated to Project 5.
b) Final platform design will be developed with Project 5 based upon existing platforms at Vanderbilt University (i.e., brain-on-a-chip). There will be one major modification: the top of the bioreactor must be removable to accommodate the unique seeding requirements of this culture. The strength of using an existing platform is that when the modification is incorporated, the system will be compatible with existing fluidics common to all platforms in the project.
We will test known teratogens (currently VPA, warfarin, and thalidomide), once the QAPP is approved. The tests will include correlating changes in culture development (by RTPCR, histology, and promoter-reporter activity) with stress responses and anticipated markers of the known adverse outcome pathways (AOP). The specific assays for AOP assessment are to be determined. We also will integrate our model with that of Project 4 (Liver):
a. Proof-of-concept experiments will involve the use of liver-conditioned medium containing liver bio-activated teratogens.
b. Physical integration of liver and limb systems will be initiated once the microfluidic platform for OCM1 and OCM2 is created. The goal of this experiment is to compare culture development under perfusion with several different media: control limb system medium (CM+ appropriate for OCM or HM+ appropriate for OCM2); limb media CM+ or MH+ with a range of teratogen doses including NOAEL, IC50, and LD50; liver medium (LM); and liver medium with a range of teratogen doses including NOAEL, IC50, and LD50. Culture health will be assessed by RT-PCR and histology.
Project 3 now has exposed a HRT-8/SV-neo trophoblast monolayer to 10 nM TCDD for 48 hours inside a single chamber IFMOC device. The next step will be to repeat these studies in the presence of a secondary inflammatory challenge with LPS. Murine studies ongoing in our lab, but not funded by the EPA, demonstrated that prior exposure to TCDD significantly enhances the maternal inflammatory response to GBS. These studies will guide our introduction of a second stressor (i.e., GBS, LPS, or second toxicant) into the IFMOC.
Project 4 will develop new biosensors and work with Projects 1-3 to incorporate mechanism-based fluorescent protein biosensors into their OCMs. Additionally, we will work with Project 5 to test our liver OCM on the integrated platform, test and optimize our liver OCM for integration with other OCMs from Projects 1-3, and validate liver readouts on the integrated platform. We also will work to validate iPSC hepatocytes for inclusion in our OCM.
Project 5 will continue refinement of device integration and molding to reduce cost for platform integration and use. In addition, the ion mobility-mass spectrometry team is standing by to receive new organ-device effluent samples for analysis and the modeling team will begin performing cluster analysis and predictive modeling of OCM responses.
Journal Articles: 57 Displayed | Download in RIS Format
| Other center views: | All 169 publications | 57 publications in selected types | All 56 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Alexander PG, Clark KL, Tuan RS. Prenatal exposure to environmental factors and congenital limb defects. Birth Defects Research, Part C: Embryo Today: Reviews 2016;108(3):243-273. |
R835736 (2015) R835736 (2016) R835736 (2017) R835736C002 (2016) R834513 (Final) |
Exit Exit Exit |
|
|
Anders AP, Gaddy JA, Doster RS, Aronoff DM. Current concepts in maternal-fetal immunology: recognition and response to microbial pathogens by decidual stromal cells. American Journal of Reproductive Immunology 2017;77(3):e12623 (14 pp.). |
R835736 (2016) |
Exit |
|
|
Argemie J, Latasa M, Atkinson S, Blokhin I, Massey V, Gue J, Cabezas J, Lozano J, Booven D, Bell A, Cao S, Vernetti L, Arab J. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nature Communications 2019;10(1):3126. |
R835736 (2018) |
Exit |
|
|
Auner AW, Tasneem KM, Markov DA, McCawley LJ, Hutson MS. Chemical-PDMS Binding Kinetics and Implications for Bioavailability in microfluidic Devices. Lab on a Chip 2019;19(5):864-874 |
R835736C001 (2018) R835736C001 (Final) R835736C005 (2018) |
Exit |
|
|
Bruner-Tran KL, Duleba AJ, Taylor HS, Osteen KG. Developmental toxicant exposure is associated with transgenerational adenomyosis in a murine model. Biology of Reproduction 2016;95(4):73 (10 pp.). |
R835736 (2015) R835736 (2016) R835736C003 (2018) |
Exit Exit |
|
|
Bruner-Tran KL, Gnecco J, Ding T, Glore DR, Pensabene V, Osteen KG. Exposure to the environmental endocrine disruptor TCDD and human reproductive dysfunction: translating lessons from murine models. Reproductive Toxicology 2017;68:59-71. |
R835736 (2015) R835736 (2016) R835736 (2017) R835736C003 (2016) R835736C003 (2017) R835736C003 (2018) R826300 (Final) |
Exit Exit Exit |
|
|
Bruner-Tran KL, Mokshagundam S, Herington JL, Osteen KG. Rodent Models of Experimental Endometriosis: Identifying Mechanisms of Disease and Therapeutic Targets. Current Women's Health Reviews 2018;14(2):173–188 |
R835736C003 (2018) |
Exit Exit |
|
|
Cyr KJ, Avaldi OM, Wikswo JP. Circadian hormone control in a human-on-a-chip:in vitro biology’s ignored component? Experimental Biology and Medicine 2017;242(17):1714-1731. |
R835736 (2017) R835736C005 (2017) |
Exit |
|
|
Ding T, Lambert LA, Aronoff DM, Osteen KG, Bruner-Tran KL. Sex-dependent influence of developmental toxicant exposure on group B Streptococcus -mediated preterm birth in a murine model. Reproductive Sciences 2018;25(5):662-673. |
R835736 (2017) R835736C003 (2017) R835736C003 (2018) |
Exit |
|
|
Dodds JN, May JC, McLean JA. Investigation of the complete suite of the leucine and isoleucine isomers: toward prediction of ion mobility separation capabilities. Analytical Chemistry 2017;89(1):952-959. |
R835736 (2015) R835736 (2016) R835736 (2017) |
Exit Exit Exit |
|
|
Ellis B, Fischer C, Martin L, Bachmann B, McLean J. Spatiochemically Profiling Microbial Interactions with Membrane Scaffolded Desorption Electrospray Ionization-Ion Mobility-Imaging Mass Spectrometry and Unsupervised Segmentation. Analytical Chemistry 2019;91(21):13703-13711. |
R835736 (2018) |
Exit |
|
|
Ellis B, Babele P, May J, Johnson C, Pfleger B, Young J, McLean J. Accelerating strain phenotyping with desorption electrospray ionization-imaging mass spectrometry and untargeted analysis of intact microbial colonies. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 2022;118(49):e2109633118. |
R835736 (Final) |
Exit Exit |
|
|
Ellis B, Babele P, May J, Johnson C, Pfleger B, Young J, McLean J. Molecular Gatekeeper Discovery:Workflow for Linking Multiple Exposure Biomarkers to Metabolomics br. ENVIRONMENTAL SCIENCE & TECHNOLOGY 2022;56(10):6162-6171. |
R835736 (Final) |
Exit Exit |
|
|
Gnecco JS, Anders AP, Cliffel D, Pensabene V, Rogers LM, Osteen K, Aronoff DM. Instrumenting a fetal membrane on a chip as emerging technology for preterm birth research. Current Pharmaceutical Design 2017;23(40):6115-6124. |
R835736 (2016) R835736 (2017) R835736C003 (2016) R835736C003 (2017) R835736C003 (2018) |
Exit Exit |
|
|
Gnecco JS, Pensabene V, Li DJ, Ding T, Hui EE, Bruner-Tran KL, Osteen KG. Compartmentalized culture of perivascular stroma and endothelial cells in a microfluidic model of the human endometrium. Annals of Biomedical Engineering 2017;45(7):1758-1769. |
R835736 (2017) R835736C003 (2016) |
Exit Exit Exit |
|
|
Gnecco JS, Pensabene V, Li DJ, Ding T, Hui EE, Bruner-Tran KL, Osteen KG. Compartmentalized culture of perivascular stroma and endothelial cells in a microfluidic model of the human endometrium. Annals of Biomedical Engineering 2017;45(7):1758-1769. |
R835736 (2015) R835736 (2016) R835736C003 (2017) R835736C003 (2018) |
Exit Exit Exit |
|
|
Gnecco JS, Ding T, Smith C, Lu J, Bruner-Tran KL, Osteen KG. Hemodynamic forces enhance decidualization via endothelial-derived prostaglandin E2 and prostacyclin in a microfluidic model of the human endometrium. Human Reproduction 2019;43(4):702-714 |
R835736C003 (2018) R839501 (2022) |
Exit |
|
|
Gough A, Vernetti L, Bergenthal L, Shun TY, Taylor DL. The MicroPhysiology Systems Database for analyzing and modeling compound interactions with human and animal organ models. Applied In Vitro Toxicology 2016;2(2):103-117. |
R835736 (2017) |
Exit Exit |
|
|
Gough A, Vernetti L, Bergenthal L, Shun TY, Taylor DL. The MicroPhysiology Systems Database for analyzing and modeling compound interactions with human and animal organ models. Applied In Vitro Toxicology 2016;2(2):103-117. |
R835736 (2015) R835736 (2016) R835736C004 (2017) |
Exit Exit |
|
|
Hawkins K, Casolaro C, Brown JA, Edwards D, WIksow J. The Microbiome and the Gut-Liver-Brain Axis for Central Nervous System Clinical Pharmacology:Challenges in Specifying and Integrating In Vitro and In Silico Models. Clinical Pharmacology & Theraputics 2020 |
R835736 (2018) |
Exit Exit |
|
|
Hutson MS, Alexander PG, Allwardt V, Aronoff DM, Bruner-Tran KL, Cliffel DE, Davidson JM, Gough A, Markov DA, McCawley LJ, McKenzie JR, McLean JA, Osteen KG, Pensabene V, Samson PC, Senutovitch NK, Sherrod SD, Shotwell MS, Taylor DL, Tetz LM, Tuan RS, Vernetti LA, Wikswo JP. Organs-on-chips as bridges for predictive toxicology. Applied In Vitro Toxicology 2016;2(2):97-102. |
R835736 (2015) R835736 (2016) R835736 (2017) R835736C001 (2018) R835736C003 (2016) R835736C003 (2017) R835736C003 (2018) |
Exit |
|
|
Hutson MS, Leung MCK, Baker NC, Spencer RM, Knudsen TB. Computational model of secondary palate fusion and disruption. Chemical Research in Toxicology 2017;30(4):965-979. |
R835736 (2015) R835736 (2016) R835736 (2017) |
Exit Exit Exit |
|
|
Iannetti L, D'Urso G, Conoscenti G, Cutri E, Tuan RS, Raimondi MT, Gottardi R, Zunino P. Distributed and Lumped Parameter Models for the Characterization of High Throughput Bioreactors. PLoS One 2016;11(9):e0162774 (25 pp.). |
R835736 (2016) |
Exit Exit |
|
|
Karolak A, Markov DA, McCawley LJ, Rejniak KA. Towards personalized computational oncology: from spatial models of tumour spheroids, to organoids, to tissues. Journal of the Royal Society Interface 2018;15(138):20170703. |
R835736C001 (2017) |
Exit Exit Exit |
|
|
Kimmel DW, Rogers LM, Aronoff DM, Cliffel DE. Prostaglandin E2 regulation of macrophage innate immunity. Chemical Research in Toxicology 2016;29(1):19-25. |
R835736 (2016) R835738 (2017) |
Exit Exit Exit |
|
|
Lee-Montiel FT, George SM, Gough AH, Sharma AD, Wu J, DeBiasio R, Vernetti LA, Taylor DL. “Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp Biol Med 2017;242(16):1617-1632. |
R835736C004 (2018) |
Exit |
|
|
Leung MC, Hutson MS, Seifert AW, Spencer RM, Knudsen TB. Computational modeling and simulation of genital tubercle development. Reproductive Toxicology 2016;64:151-161. |
R835736 (2015) R835736 (2016) R835736C005 (2016) |
Exit Exit Exit |
|
|
Leung MCK, Hutson MS, Seifert AW, Spencer RM, Knudsen TB. Computational modeling and simulation of genital tubercle development. Reproductive Toxicology 2016;64:151-161. |
R835736 (2017) |
Exit Exit Exit |
|
|
Li X, George SM, Vernetti L, Gough AH, Taylor DL. A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab on a Chip 2018;21;18(17):2614-2631 |
R835736C004 (2018) |
Exit |
|
|
May JC, Morris CB, McLean JA. Ion mobility collision cross section compendium. Analytical Chemistry 2017;89(2):1032-1044. |
R835736 (2016) |
Exit |
|
|
May J, Jurneczko E, Stow A, Kratochvil I, Kalhof S, McLean J. Conformational landscapes of ubiquitin, cytochrome c, and myoglobin: Uniform field ion mobility measurements in helium and nitrogen drift gas. INTERNATIONAL JOURNAL OF MASS SPECTROMETRY 2018;427:79-90 |
R835736 (Final) |
Exit |
|
|
Melow S, Miller D, Gizzie E, Cliffel D. A low-interference, high-resolution multianalyte electrochemical biosensor. Analytical Methods 2020;12(31):3873-3882. |
R835736 (Final) |
Exit |
|
|
MHarris R, May J, Stinson C, Xia Y, McLean J. Determining Double Bond Position in Lipids Using Online Ozonolysis Coupled to Liquid Chromatography and Ion Mobility-Mass Spectrometry. ANALYTICAL CHEMISTRY 2018;90(3):1959-1924 |
R835736 (Final) |
Exit |
|
|
Morris C, May J, Leaptrot K, McLean J. Evaluating Separation Selectivity and Collision Cross Section Measurement Reproducibility in Helium, Nitrogen, Argon, and Carbon Dioxide Drift Gases for Drift Tube Ion Mobility-Mass Spectrometry. JOURNAL OF THE AMERICAN SOCIETY FO MASS SPECTROMETRY 2019;30(6):1059-1068 |
R835736 (Final) |
Exit |
|
|
Nichols C, May J, Sherrod S, McLead J. Automated flow injection method for the high precision determination of drift tube ion mobility collision cross sections. ANALYST 2018;143(7):1556-1559 |
R835736 (Final) |
Exit |
|
|
Nichols C, Dodds J, Rose B, Piache J, Morris C, Codreau S, May J, Sherrod S, McLean J. Untargeted Molecular Discovery in Primary Metabolism: Collision Cross Section as a Molecular Descriptor in Ion Mobility-Mass Spectrometry. ANALYTICAL CHEMISTRY 2018;90(24):14484-14492 |
R835736 (Final) |
Exit |
|
|
Richardson L, Gnecco J, Ding T, Osteen K, Rogers L, Arnoff DM, Menon R. Fetal Membrane Organ-On-Chip:An Innovative Approach to Study Cellular Interactions. Reproductive Sciences 2020;. |
R835736 (2018) |
Exit |
|
|
Rogers JM, Anders AP, Doster RS, Gill EA, Gnecco JS, Holley JM, Randis TM, Ratner AJ, Gaddy JA, Osteen K, Arnoff DM. Decidual stromal cell-derived PGE(2) regulates macrophage responses to microbial threat. American Journal of Reproductive Immunology 2018;80(4). |
R835736C003 (2018) |
Exit |
|
|
Sakolish C, Luo Y, Valdiviezo A, Vernetti L, Rusyn I, Chiu W. Prediction of hepatic drug clearance with a human microfluidic four-cell liver acinus microphysiology system. Toxicology 2021;463. |
R835736 (Final) |
Exit Exit |
|
|
Sakolish C, Reese CE, Luo YS, Valdiviezo A, Schurdak ME, Gough A, Taylor DL, Chiu WA, Venetti LA, Rusyn I. Analysis of reproducibility and robustness of a human microfluidic four-cell liver acinus microphysiology system (LAMPS). Toxicology 2021;448. |
R835736 (Final) R840032 (2021) |
Exit Exit |
|
|
Schurdak M, Vernetti L, Bergenthal L, Wolter Q, Shun T, Karcher S, Taylor D, Gough A. Applications of the microphysiology systems database for experimental ADME-Tox and disease models. LAB ON A CHIP 2020;20(8):1472-1492. |
R835736 (2018) |
Exit |
|
|
Senutovitch N, Vernetti L, Boltz R, DeBiasio R, Gough A, Taylor DL. Fluorescent protein biosensors applied to microphysiological systems. Experimental Biology and Medicine 2015;240(6):795-808. |
R835736 (2015) R835736 (2016) R835736C004 (2015) R835736C004 (2016) |
Exit |
|
|
Soto-Gutierrez A, Gough A, Vernetti LA, Taylor DL, Monga SP. Pre-clinical and clinical investigations of metabolic zonation in liver diseases: the potential of microphysiology systems. Experimental Biology and Medicine 2017;242(16):1605-1616. |
R835736 (2017) R835736C004 (2017) |
Exit |
|
|
Stocks MM, Crispens MA, Ding T, Mokshagundam S, Bruner-Tran KL, Osteen KG. Therapeutically targeting the inflammasome product in a chimeric model of endometriosis-related surgical adhesions. Reproductive Sciences 2017;24(8):1121-1128. |
R835736 (2015) R835736 (2016) |
Exit |
|
|
Stow S, Onifer T, Forsythe J, Jefzger H, Keiecien N, May J, McLean J, Hercules D. Structural Characterization of Methylenedianiline Regioisomers by Ion Mobility-Mass Spectrometry and Tandem Mass Spectrometry. 4. 3-Ring and 4-Ring Isomers. ANALYTICAL CHEMISTRY 2015;87(12):6288-6296 |
R835736 (Final) |
Exit |
|
|
Stow S, Causon T, Zheng X, Krulugama R, Meringer T, May J, Rennie E, Baker E, Smith R, McLean J, Hann S, Fjeldsted J. An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. ANALYTICAL CHEMISTRY 2017;89(17):9048-9055 |
R835736 (Final) |
Exit |
|
|
Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Shun TY, Gough A, Taylor DL. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Experimental Biology and Medicine 2016;241(1):101-114. |
R835736 (2015) R835736 (2016) R835736 (2017) R835736C004 (2015) R835736C004 (2016) R835736C004 (2017) R835736C004 (2018) |
Exit |
|
|
Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Scientific Reports 2017;7:42296 (14 pp.). |
R835736 (2017) R835736C004 (2018) R835738C002 (2017) R835738C005 (2017) |
Exit Exit Exit |
|
|
Vernetti LA, Vogt A, Gough A, Taylor DL. Evolution of experimental models of the liver to predict human drug hepatotoxicity and efficacy. Clinics in Liver Disease 2017;21(1):197-214. |
R835736 (2015) R835736 (2016) R835736 (2017) R835736C004 (2017) R835736C004 (2018) |
Exit Exit |
|
|
Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Scientific Reports 2017;7:42296 (15 pp.). |
R835736 (2015) R835736 (2016) R835736C004 (2016) R835736C005 (2016) R835738 (2016) R835738C003 (2017) R835738C005 (2017) |
Exit Exit Exit |
|
|
Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Scientific Reports 2017;7:42296 (14 pp.). |
R835736C004 (2017) R835738 (2017) R835738C002 (2016) |
Exit Exit Exit |
|
|
Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Scientific Reports 2017;7:42296 (15 pp.). |
R835736 (2015) R835736 (2016) R835736C004 (2016) R835736C005 (2016) R835738 (2016) R835738C003 (2017) R835738C005 (2017) |
Exit Exit Exit |
|
|
Watson DE, Hunziker R, Wikswo JP. Fitting tissue chips and microphysiological systems into the grand scheme of medicine, biology, pharmacology, and toxicology. Experimental Biology and Medicine 2017;242(16):1559-1572. |
R835736 (2017) R835736C005 (2017) |
Exit |
|
|
Wikswo JP. Looking to the future of organs-on-chips: interview with Professor John Wikswo. Future science OA 2017;3(2):FSO163. |
R835736C005 (2017) |
Exit Exit Exit |
|
|
Miller DR, McClain ES, Cliffel DE. Electrochemical Microphysiometry Detects Cellular Glutamate Up-take. Journal of The Electrochemical Society 2018;165(12):G3120-G3124. |
R835736 (Final) R835736C005 (2018) R835736C005 (Final) |
Exit Exit Exit |
|
|
Rogers M, Sobolik T, Schaffer DK, Samson PC, Johnson AC, Owens P, Codreanu SG, Sherrod SD, McLean JA, Wikswo JP, Richmond A. Engineered microfluidic bioreactor for examining the three-dimensional breast tumor microenvironment Biomicrofluidics 2018;12(3):034102. |
R835736C005 (Final) |
Exit Exit |
|
|
Ding T et al, 2018. Reproductive Sciences 25(5):662-673 Bruner-Tran KL et al, 2018. Current Women’s Health Rev Jun;14(2):173-188 Rogers et al, 2018. American Journal of Reproductive Immunology Oct;80(4):e13032. |
R835736C003 (2018) |
not available |
Supplemental Keywords:
mammary development, mammary toxicology, organs on chip, mammary on chip, PDMS bioreactors, thick tissue bioreactor, PDMS interactions, organotypic cell culture, limb development, organotypic culture model, high density, micromass culture, adult human mesenchymal stem cell, MSC, chondrogenesis, hypertrophy, joint segmentation, three dimensional limb bioreactor, mechanical stimulation, toxicology, valproic acid, warfarin, mifepristone, microfluidic pumps and valves, integrated organ microfluidicsRelevant Websites:
Microphysiology Systems Database | University of Pittsburgh Drug Discovery Institute ExitCenter for Cellular & Molecular Engineering | University of Pittsburgh Exit
Vanderbilt Institute for Integrative Biosystems Research and Education (VIIBRE) Exit
Vanderbuilt Center for Innovative Technology Exit
Vanderbilt-Pittsburgh Resource for Organotypic Models for Predictive Toxicology (VPROMPT) Exit
Progress and Final Reports:
Original Abstract Subprojects under this Center: (EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
R835736C001 Mammosphere Bioreactor For Life-Stage Specific Toxicology
R835736C002 Organotypic Culture Model to Analyze DevelopmentalLimbMalformationsResulting from Toxicant/Teratogen Exposure
R835736C003 Validating a fetal membrane on a chip model for characterizing
reproductive toxicant exposure risks
R835736C004 Organotypic Liver Model for Predictive Human Toxicology and Metabolism
R835736C005 Systems Engineering & Analysis for Organotypic Culture Models
The perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.