Grantee Research Project Results
2017 Progress Report: Organotypic Model of Human Kidney as a Platform for Adverse Outcomes Pathway Assessment of Engineered Nanomaterials
EPA Grant Number: R835738C002Subproject: this is subproject number 002 , established and managed by the Center Director under grant R835738
(EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
Center: Predictive Toxicology Center for Organotypic Cultures and Assessment of AOPs for Engineered Nanomaterials
Center Director: Faustman, Elaine
Title: Organotypic Model of Human Kidney as a Platform for Adverse Outcomes Pathway Assessment of Engineered Nanomaterials
Investigators: Kelly, Edward J
Current Investigators: Kelly, Edward J.
Institution: University of Washington
EPA Project Officer: Aja, Hayley
Project Period: December 1, 2014 through November 30, 2018 (Extended to November 30, 2019)
Project Period Covered by this Report: December 1, 2016 through November 30,2017
RFA: Organotypic Culture Models for Predictive Toxicology Center (2013) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
a. Specific Aims
One of the primary objectives of our project is to design, implement, and test a tissue-engineered human kidney microphysiological system and to evaluate the response of exposure to engineered nanomaterials (ENMs). To this end, we evaluated the toxicological effects of two forms of silver nanoparticles (AgNPs) with an organotypic microfluidic device that utilizes the Nortis™ microphysiological (MPS) system which accurately reflects human renal physiology with the culturing of primary human proximal tubule epithelial cells (PTEC) in a physiologically relevant 3-D configuration and an appropriately scaled lumenal flow rate.
For this project we will focus on the ENMs (Quantum Dots and AgNPs) that have practical applications in photonics, optics, solar cells, sensors, imaging, and anti-microbial activity.
Our hypotheses for this project are:
1) Differentiation status of organotypic kidney cultures will define the response to ENMs.
2) The oxidative stress and endoplasmic reticulum (ER) stress responses within organotypic kidney cultures will be key common response pathways associated with exposure to ENMs.
3) Inflammatory pathways within organotypic kidney cultures will have significant and specific impacts beyond general oxidative stress.
4) Genetic differences among organotypic kidney cultures will be critical factors in interpreting kidney specific responses.
5) Using a systems-based Adverse Outcome Pathway (AOP) analysis with toxicokinetic and dynamic models as a part of a risk assessment framework will allow for cross assay and organ interpretation.
In order to address these hypotheses, this project will focus on developing an organotypic kidney model systems derived from human kidney. We will use a microphysiometer flow through system developed by Nortis Inc, to establish and differentiate long-term cultures of kidney proximal tubule cells for this project. Accordingly, our Specific Aims are:
Specific Aim 1. Culture human kidney proximal tubular cells within Nortis microphysiometers, and determine optimum conditions for retaining long-term viability and function. Viability and function will be assessed using fluorescence probes, enzyme assays, mRNA and protein expression analysis and model kidney toxicants.
Specific Aim 2. Assess the in vitro uptake and toxicity of QDs and AgNPs in these 3 microphysiometer models of kidney. This will be done under both short (1 day) and long term (3-4 weeks) exposure scenarios, meant to model acute and repeated exposure to these ENMs. We will examine the effects of these ENMs on cell viability, oxidative stress, ER-based stress, and inflammation pathways within and across the different species being tested.
Information gathered from these studies will be valuable in helping to predict the role of ENM constituent metals on their toxicity, as well as in the design of cores and coatings that will optimize the desirable properties of these materials while minimizing their adverse health impacts. Ultimately, these data will be useful in risk assessments of these materials in susceptible humans, and thus to inform policy-making.
Progress Summary:
Assessment of Nephrotoxicity with Engineered Nanomaterials
Our nephrotoxicity investigations with ENM and our Nortis MPS have been hampered by the general insolubility of most ENM in our in vitro system which utilizes serum-free media, i.e, quantum dots with a Cd and Se core and silver nanoparticles (AgNPs) that we have considered for evaluation. Since our the protein coat (corona), which forms on the surface of the ENM solutions when dissolved in serum-containing medias, does not occur therefore increasing the chances of aggregation and precipitation. Previous investigations with Ag nanoparticles were confounded by solubility issues and cell culture media modifications to make the media more amenable to solubility contributed to the observed toxicity.
Quantum Dots
Quantum dots (QD) with a CdSe/ZnS core and a net positive charge [polydiallydimethylammonium chloride (PDDA) coating from Ocean NanoTech ID QSQ-600-02] were chosen for PTEC exposure experiments due to their nephrotoxic potential by having a cadmium/selenium core and for their favorable solubility capabilities. Four independent kidney toxicity experiments were completed with QD that have the CdSe/ZnS core. The positive control condition for these experiments was 25 mM CdCl2. All treatments were 48 hour exposures with MPS effluent samples collected at 24 and 48 hours for injury biomarkers, miRNA and Cd dosimetry. PTECs were seeded into a lumen formed in the collagen bed in the MPS and cultured under flow (0.5 mL/min) for at least five days before treatment. Endpoint evaluations included: RNA transcript analysis, chip effluent biomarker analysis for kidney injury markers- KIM-1, calbindin, clusterin, osteoactivin, TFF3, VEGF-A, and αGST. Syringe and effluent cadmium concentrations were evaluated by inductively coupled plasma mass spectrometry (ICP-MS) to ascertain exposure concentrations. ICP-MS of QD stock solutions was performed to assess free cadmium concentrations.
Two QD dose-response toxicity experiments were performed with PDDA QDs with the first experiment evaluating QD concentrations at 0.025, 0.25 and 2.5 nM. Device effluents were evaluated for KIM-1, calbindin, clusterin, osteoactivin, TFF3, VEGF-A, and αGST using the MesoScale Discovery Kidney Injury Panel 3 kit that uses electrochemiluminescence technology. Kidney injury panel results from this dose-response experiment did not show a clear dose-response, including KIM-1, with high variability especially in the negative controls making data interpretation difficult. We will most likely not be using this kit again for our work.
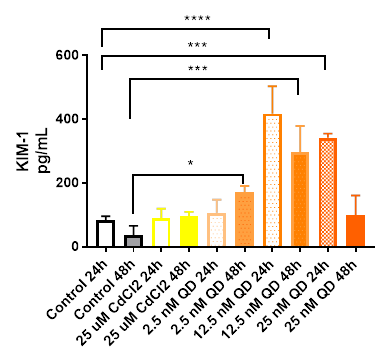
A second 48 hour dose-response experiment with QD was conducted at concentrations of 2.5, 12.5 and 25 nM with 25 mM CdCl2 again used as the positive control. With these higher concentrations, a clear dose-response was observed with significant increases in KIM-1 concentrations in the MPS effluents with QD concentrations (Figure 1). In addition, the effluents were evaluated for miRNA analyses (courtesy of Susanne Ramm- Harvard Medical School) for transcripts previously associated with renal injury. Figure 2 illustrates changes in miRNA transcripts after 48h exposure to 25 mM CdCl2 from this experiment (the positive control chips) and an experiment completed last year with the same treatment conditions but different PTEC donor. Treatment-related (25 mM CdCl2) changes in both miRNA data sets were modest. Following the QD exposure period, cells were flushed out of some of the devices with a detergent and, following the addition of Trizol and purification by the Qiagen mRNeasy kit, were submitted for RNAseq analysis (results pending). The QD stock solution (8 mM) was analyzed for cadmium concentration using ICP/MS to ascertain if free cadmium could be detected. The measured cadmium concentration was 349 ng/mL suggesting that free cadmium exposure can occur with PDDA QD exposure. The working QD concentrations used in our experiments (from 0.25 and 25 nM) which corresponds to 0.011 and 1.1 ng/mL of free cadmium, respectively and with the reporting limit for Cd by ICP/MS set at 1 ng/mL there was, as expected, no measurable cadmium detected in effluents of MPS chips exposed to 2.5 nM of PDDA QD.
Figure 1. MPS effluents were collected at 24 and 48 hours and analyzed for human KIM-1. KIM-1 concentrations in the effluents of PTECs exposed to QDs increased in * p<0.05; *** p= 0.0004: ****p=0.0001.
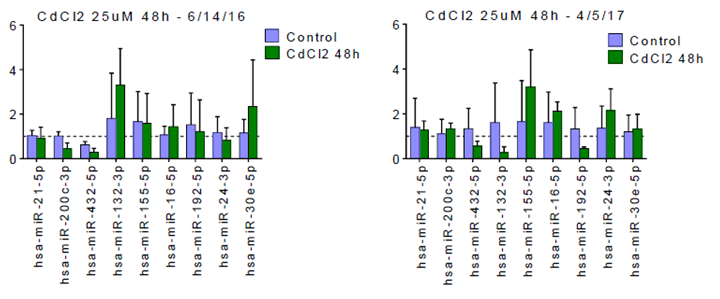
Figure 2. miRNA abundance (represented in fold-change in chip effluents) for transcripts that have been associated with kidney injury and/or inflammation. The data are from two independent experiments with PTECs from two different donors. The panel on the right represents data from the positive control treatment (25 m M CdCl2) for the second QD dose-response experiment. The panel on the left is miRNA data from a positive control treatment performed a year earlier using cells from a different donor.
Cadmium Chloride
Since cadmium nephrotoxicity is a primary concern for systemic exposures to QDs with a Cd/Se core we continued exploring AOPs for cadmium exposure in kidneys. In 2017, we exposed PTECs from six different donors to a range of Cd Cl2 concentrations (2.5-25 mM) to determine dose-responses at the RNA transcript level, ICC staining, dosimetry analyses and probed effluents for kidney injury markers. To bolster our understanding of cadmium nephrotoxicity effects, we evaluated RNA transcript data from dose-response experiments with CdCl2 conducted in 2016 and performed cadmium dosimetry by ICP/MS in the dosing syringes and MPS effluents to obtain a better understanding of PTEC exposures and material (cadmium) loss throughout the system. We later coupled a liver MPS containing human primary hepatocytes with our standard PTEC MPS to determine if there were hepatic factors contributing to cadmium nephrotoxicity as cadmium is also toxic to the liver.
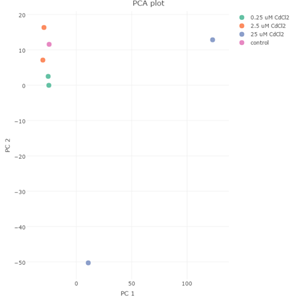
We exposed PTECs to 0, 0.25, 2.5 and 25 mM CdCl2 for 48 hours and isolated RNA from the cells in the MPS after the exposure period with 25 mM being the EC50 concentration and the designated positive control condition for the bulk of our work. After exposures, we isolated RNA from two chips at each concentration and had transcripts analyzed by RNAseq. Principal components analysis (PCA) of the transcripts of the four CdCl2 treatment groups reveals that effects of CdCl2 treatment on PTEC transcript levels occurs at concentrations greater than 2.5 mM therefore for analysis purposes we pooled the RNA transcript responses for the 0, 0.25 and 2.5 mM groups and compared that with transcript changes from the 25 mM CdCl2 group (Figure 3).
Figure 3. Principal component analysis of the RNA transcript data for the 0, 0.25, 2.5 and 25 m M CdCl2-treated PTECs shows that the 25 m M CdCl2 dose group is separated from the other groups suggesting that the changes in RNA transcripts occurs at CdCl2 concentrations greater than 2.5 m M and therefore we pooled the data for the 0, 0.25 and 2.5 mM groups in comparison to the 25 m M group.
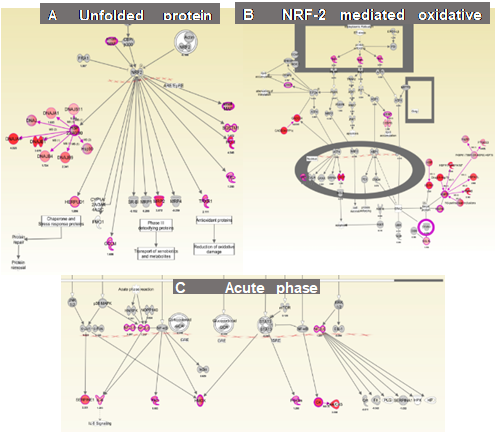
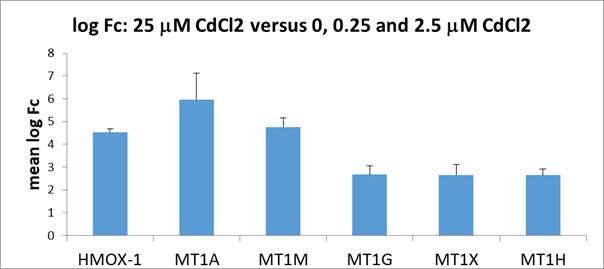
RNA seq transcript analyses of the CdCl2-treated PTECs RNA transcript analyses of PTECS exposed to 25 mM CdCl2 for 48 hours has shown an increase in mRNA transcripts, relative to controls (i.e., 0, 0.25 and 2.5 mM CdCl2). A list of the top twenty upregulated RNA transcripts based on logFC scores is shown in Figure 4. Our false discovery rate (FDR) cutoff was 5% and we selected genes based on a 1.5-fold or greater difference in expression. Pathway analyses of RNA transcript changes with 25 mM CdCl2 versus 0, 0.25 and 2.5 mM by Ingenuity Pathway Analysis (IPA) determined that the following pathways are involved with Cd exposure: A) Unfolded protein response; B) NRF-2 mediated oxidative stress response and C) Acute phase response
(Figure 5) and several metallothionein members (MT1A, MT1M, MT1G, MT1X and MT1H) (Figure 6) which are consistent with known cellular responses of a protective mechanism from cadmium exposure.
| SYMBOL | GENENAME | logFC | logCPM | FDR |
|---|---|---|---|---|
| HSPA6 | heat shock protein family A (Hsp70) member 6 | 7.68 | 9.87 | 0.00 |
| HSPA1B | heat shock protein family A (Hsp70) member 1B | 6.22 | 14.48 | 0.05 |
| LINC00624 | long intergenic non-protein coding RNA 624 | 5.44 | 2.57 | 0.00 |
| HSPA1A | heat chock protein family A (Hsp70) member 1A | 5.31 | 12.40 | 0.03 |
| ANKRD1 | enkyrin repeat domain 1 | 5222 | 6.61 | 0.00 |
| MTM1M | metallothionein 1M | 5.11 | 6.94 | 0.00 |
| MT1A | metallothionein 1A | 4.94 | 3.77 | 0.02 |
| MAFA | MAF bZIP transcription factor A | 4.90 | 2.67 | 0.01 |
| GADD45G | growth arrest and DNA damage inducible gamma | 4.76 | 2.51 | 0.01 |
| VGF | VGF nerve growth factor inductible | 4.62 | 5.12 | 0.03 |
| HMOX1 | heme oxygenase 1 | 4.40 | 8.45 | 0.00 |
| DNAJA4 | DnaJ heat shock protein family (Hsp40) member A4 | 4.39 | 7.81 | 0.00 |
| ACTN2 | actinin alpha 2 | 4.36 | 3.22 | 0.03 |
| HS3ST2 | heparan sulfate-glucosamine 3-sulfotransferase 2 | 4.31 | 2.08 | 0.03 |
| FOSB | FosB proto-oncogene, AP-1 transcription factor sunbunit | 4.26 | 3.85 | 0.01 |
| IL11 | interleukin 11 | 3.93 | 4.25 | 0.00 |
| PPP1R15A | protein phosphatase 1 regulatory subunit 15A | 3.91 | 8.25 | 0.01 |
| LUCAT1 | lung cancer associated transcript 1 (non-protein coding) | 3.89 | 1.88 | 0.03 |
| INHBA | inhibin beta A subunit | 3.86 | 3.06 | 0.01 |
Figure 4. A list of the top twenty up-regulated (logFC) transcripts includes several metallothionein genes (MT1M and MT1A), heme oxygenase (used as an oxidative stress marker) and several heat shock proteins that are part of the unfolded-protein stress response.
Figure 5. Shown are three canonical pathways generated by IPA of RNA transcript changes using RNAseq in PTECs exposed to CdCl2 for 48 hours when compared to 0, 0.025 and 2.5 m M CdCl2. Numbers near each transcript ID indicates fold-change increases compared to controls.
IPA has determined that CdCl2 exposure has upregulated many genes in the A) Unfolded protein response; B) NRF-2 mediated oxidative stress response and C) Acute phase response.
Figure 6. Fold-change differences (log Fc) in mRNA from PTECs exposed to 25 uM CdCl2 for 48 hours. Increase transcript levels were seen with: heme oxygenase 1 (HMOX-1) and five members of metallothioneins (MT1A, MT1M, MT1G, MT1X and MT1H). Error bars indicate standard deviations.
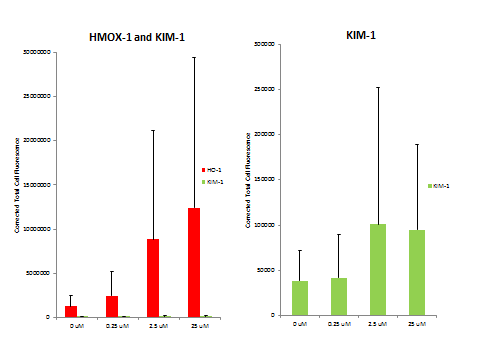
We also analyzed some MPS devices from the dose-response experiments by immunocytochemistry for heme oxygenase 1 and KIM-1 (Figure 7). There was a dose-responsive increase in HMOX-1 and KIM-1 as measured by immunofluorescence (Figure 8).
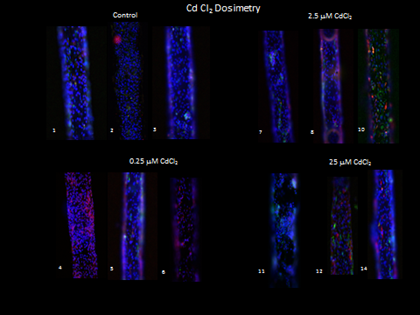
Figure 7. Immunocytochemistry staining for heme oxygenase 1 (HMOX-1: red) and KIM-1 (green) increased in staining for both proteins with increased Cd dose.
Figure 8. Corrected total cell fluorescence of the HMOX-1 and KIM-1 signal in dose-response 48 hour exposure to CdCl2.
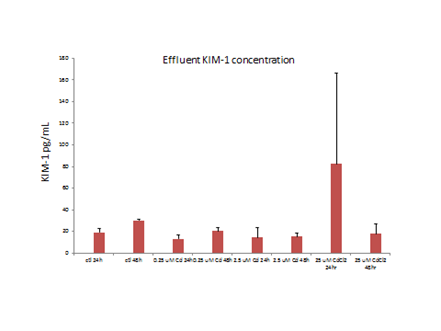
Analyses of MPS effluents of PTECs exposed to ≥2.5 uM CdCl2 had increased KIM-1 concentrations, relative to controls (Figure 9).
Figure 9. Effluent concentrations of KIM-1increased with dose over 2.5 m M CdCl2.
Dosimetry
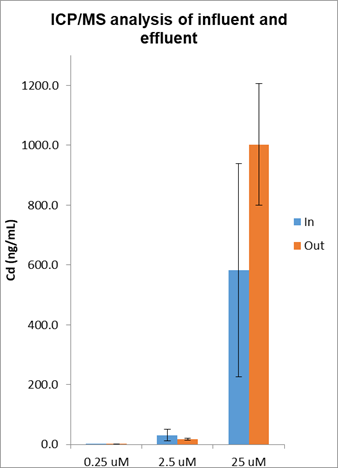
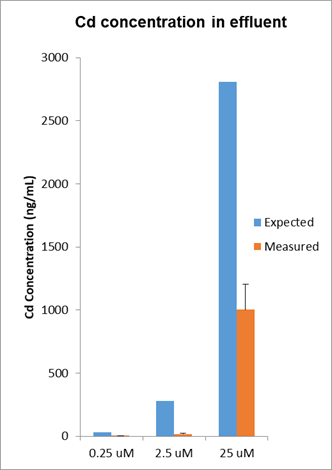
We addressed the fundamental question of understanding the actual cadmium exposure during the course of several MPS experiments by sampling the dosing solutions contained in the syringe and taking a sample of effluent to get a handle of material loss through the system (Figure 10). Cadmium is exposed to at least seven different materials within an MPS device including the tubing and syringe assembly while also exposed to 5,000-10,000 cells that are either live and intact, dead and lysed or somewhere in between. Cadmium concentrations decreased by ~75% based on expected and measured Cd concentrations of the effluents of the MPS.
Figure 10. Cadmium dosimetry by ICP/MS was used to determine cadmium exposure levels during the experiments that yielded the RNA-seq data presented in Figure 5. Results indicate that the cadmium concentrations measured in the influent and effluent were variable and in general the concentrations in the effluents were considerably less than what was expected suggesting that cadmium was accumulating in the MPS system.
Liver:kidney coupled evaluation of CdCl2
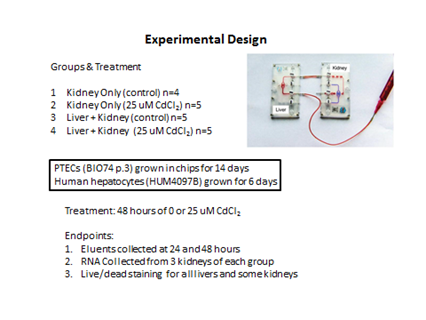
To address whether there are factors released by cadmium exposed hepatocytes that could influence renal toxicity, we coupled a liver MPS containing human primary hepatocytes with a kidney chip containing human PTECs and placed the coupled system under flow as illustrated in Figure 11.
Figure 11. Experimental design of the initial liver:kidney coupled experiment exposed to 25 m M CdCl2 under 30 m L/hr flow.
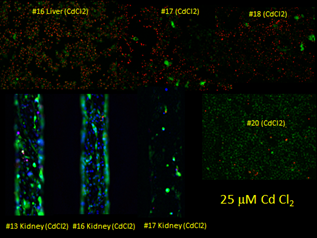
Our initial experiment of the liver:kidney coupled experiment at 25 mM CdCl2 proved to be too high of a concentration as most of the hepatocytes were dead after 48 hours (Figure 12). We plan to repeat the experiments at 1 and 5 mM CdCl2 and evaluate kidney effluents and RNA transcripts.
Figure 12. Live/dead staining of control and 25 m M CdCl2-treated liver and kidney chips. Live (green) stain is calcein AM while the dead stain (red) is ethidium homodimer-1and the nuclear stain (blue) is NucBlue™ from Molecular Probes.
Future Activities:
To address whether there are factors released by cadmium-exposed hepatocytes that could influence renal toxicity, we have started to couple liver MPS devices containing human primary hepatocytes with an MPS PTEC device and placed the coupled system under flow. We will be comparing renal toxicity with or without a coupled liver and evaluate mRNA transcripts and biomarkers from the renal MPS to ascertain mechanistic changes. We will continue to evaluate QD renal toxicity with comparisons to cadmium renal toxicity as a reference.
We are evaluating RNA transcript changes in PTECs exposed to CdSe/ZnS core/shell quantum dots. We have demonstrated that we can detect modest PTEC toxicity with increased HO-1 and KIM-1 immunofluorescence in MPS after 48 hour exposure to 2.5 and 25 nM QDs and comparisons to the CdCl2 toxicity pathways is forthcoming. We plan to pursue experiments that involve coupling of the liver MPS with the kidney chip and explore interactions between the organ systems during CdCl2 and QD exposures. Adverse Outcome Pathway (AOP) determinations using RNA-seq transcriptomic analysis and immunocytochemistry (ICC) fluorescent imaging for KIM-1 and HO-1 and MPS effluent analysis for KIM-1 and ICP/MS measurements of effluents will continue. Other response ICC markers that will be evaluated include: SGLT2 and E-cadherin while additional effluent markers include interleukin 18 and Neutrophil gelatinase-associated lipocalin (NGAL) and any promising biomarkers noted in our RNAseq analyses.
Journal Articles on this Report : 9 Displayed | Download in RIS Format
| Other subproject views: | All 64 publications | 17 publications in selected types | All 16 journal articles |
|---|---|---|---|
| Other center views: | All 150 publications | 50 publications in selected types | All 49 journal articles |
| Type | Citation | ||
|---|---|---|---|
|
|
Chang S-Y, Weber EJ, Sidorenko VS, Chapron A, Yeung CK, Gao C, Mao Q, Shen D, Wang J, Rosenquist TA, Dickman KG, Neumann T, Grollman AP, Kelly EJ, Himmelfarb J, Eaton DL. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2017;2(22):95978 (15 pp.). |
R835738 (2017) R835738C002 (2017) R835738C002 (2018) |
Exit Exit Exit |
|
|
Chang S-Y, Voellinger JL, Van Ness KP, Chapron B, Shaffer RM, Neumann T, White CC, Kavanagh TJ, Kelly EJ, Eaton DL. Characterization of rat or human hepatocytes cultured in microphysiological systems (MPS) to identify hepatotoxicity. Toxicology In Vitro 2017;40:170-183. |
R835738 (2016) R835738 (2017) R835738C002 (2016) R835738C002 (2017) R835738C003 (2016) R835738C003 (2017) |
Exit Exit Exit |
|
|
Nolin JD, Lai Y, Ogden HL, Manicone AM, Murphy RC, An D, Frevert CW, Ghomashchi F, Naika GS, Gelb MH, Gauvreau GM, Piliponsky AM, Altemeier WA, Hallstrand TS. Secreted PLA2 group X orchestrates innate and adaptive immune responses to inhaled allergen. JCI Insight 2017;2(21):e94929 (18 pp.). |
R835738 (2017) R835738C001 (2018) R835738C002 (2017) |
Exit Exit Exit |
|
|
Scoville DK, Botta D, Galdanes K, Schmuck SC, White CC, Stapleton PL, Bammler TK, MacDonald JW, Altemeier WA, Hernandez M, Kleeberger SR, Chen LC, Gordon T, Kavanagh TJ. Genetic determinants of susceptibility to silver nanoparticle-induced acute lung inflammation in mice. FASEB Journal 2017;31(10):4600-4611. |
R835738 (2017) R835738C001 (2017) R835738C001 (2018) R835738C002 (2017) |
Exit |
|
|
Van Ness KP, Chang SY, Weber EJ, Zumpano D, Eaton DL, Kelly EJ. Microphysiological systems to assess nonclinical toxicity. Current Protocols in Toxicology 2017;73(1):14.18.1-14.18.28. |
R835738 (2017) R835738C001 (2018) R835738C002 (2017) R835738C002 (2018) R835738C004 (2018) R835738C005 (2017) |
Exit |
|
|
Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Scientific Reports 2017;7:42296 (14 pp.). |
R835738C002 (2017) R835738C005 (2017) R835736 (2017) R835736C004 (2018) |
Exit Exit Exit |
|
|
Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Corrigendum: Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Scientific Reports 2017;7:44517. |
R835738 (2016) R835738 (2017) R835738C002 (2016) R835738C002 (2017) R835738C005 (2017) |
Exit |
|
|
Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, Wang Z, Yamaura Y, Hailey DW, Neumann T, Shen DD, Thummel KE, Muczynski KA, Himmelfarb J, Kelly EJ. Development of a microphysiological model of human kidney proximal tubule function. Kidney International 2016;90(3):627-637. |
R835738 (2016) R835738 (2017) R835738C002 (2016) R835738C002 (2017) |
Exit |
|
|
Weber EJ, Himmelfarb J, Kelly EJ. Concise review: current and emerging biomarkers of nephrotoxicity. Current Opinion in Toxicology 2017;4:16-21. |
R835738 (2017) R835738C002 (2017) R835738C002 (2018) |
Exit |
Supplemental Keywords:
Kidney, Engineered nanomaterials, Quantum Dots, Cadmium, Kidney Injury, KIM-I (Kidney Injury Molecule)Relevant Websites:
Predictive Toxicology Center Exit
Progress and Final Reports:
Original AbstractMain Center Abstract and Reports:
R835738 Predictive Toxicology Center for Organotypic Cultures and Assessment of AOPs for Engineered Nanomaterials Subprojects under this Center: (EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
R835738C001 Airway Epithelium Organotypic Culture as a Platform forAdverseOutcomesPathway Assessment of Engineered Nanomaterials
R835738C002 Organotypic Model of Human Kidney as a Platform for Adverse Outcomes
Pathway Assessment of Engineered Nanomaterials
R835738C003 Organotypic Models of Mammalian Liver as a Platform for Adverse Outcomes
Pathway Assessment of Engineered Nanomaterials
R835738C004 Organotypic Model of Testis as a Platform for Adverse Outcomes Pathway
Assessment of Engineered Nanomaterials
R835738C005 Integrating Liver, Kidney and Testis Nanomaterial Toxicity using the
Adverse Outcome Pathway Approach
The perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
Project Research Results
16 journal articles for this subproject
Main Center: R835738
150 publications for this center
49 journal articles for this center