Grantee Research Project Results
2012 Progress Report: Environmental Research and Technology Transfer Program of The Consortium for Plant Biotechnology Research, Inc.
EPA Grant Number: EM834388Center: The Consortium for Plant Biotechnology Research, Inc., Environmental Research and Technology Transfer Program
Center Director: Schumacher, Dorin
Title: Environmental Research and Technology Transfer Program of The Consortium for Plant Biotechnology Research, Inc.
Investigators: Bonning, Bryony C. , Paterson, Andrew , Cheng, Zong-Ming , Pantalone, Vincent R , Qu, Rongda , Ren, Shuxin , Yang, Guochen , Kessler, Michael
Current Investigators: Schumacher, Dorin , Cheng, Zong-Ming , Kessler, Michael , Larock, Richard C. , Paterson, Andrew , Pullman, Gerald , May, Sheldon
Institution: Iowa State University , University of Georgia , University of Tennessee , North Carolina State University
Current Institution: The Consortium for Plant Biotechnology Research, Inc
EPA Project Officer: Packard, Benjamin H
Project Period: January 1, 2009 through December 31, 2012 (Extended to December 31, 2013)
Project Period Covered by this Report: January 1, 2012 through December 31,2012
Project Amount: $1,706,000
RFA: Targeted Research Center (2009) RFA Text | Recipients Lists
Research Category: Targeted Research , Consortium for Plant Biotechnology
Objective:
Iowa State University- Bryony C. Bonning
Objective of Research: The overall goal of this project was determine whether an aphid gut binding peptide such as GBP3.1 can be combined with the Bt-derived cytolytic toxin for production of aphicidal toxins. Our ultimate goal is to produce transgenic plants that are aphid resistant. The objectives of this research project were to (i) determine which peptide characteristics are essential for binding to the aphid gut, and (ii) insert the gut binding residues into a Bt-derived cytolytic toxin, and test the modified toxins for stability in and increased binding to the aphid gut, and for toxicity. We have successfully completed both project objectives and as a consequence have developed an effective novel approach for management of aphid pests.
University of Tennessee - Vincent Pantalone
The long-term objective of this research is to develop a commercially acceptable, environmentally superior, high yielding soybean variety with low seed phytate. We have four specific aims:
- Production of low phytate soybean seeds to enable multiple environment field research and pilot plant research processing of soy meal.
- Analytical testing of the improved low phytate soybean seeds.
- Chicken feeding trials to document enhanced nutrition.
- Insertion of a novel industry proprietary RFO transgene to soybean that modifies expression as an alternative strategy to improve livestock nutrition and metabolic growth rates.
North Carolina State University - Rongda Qu
Objectives:
- Test AtAVP1, and maize CBP (calcium binding domain of careticulin) transgenic rice lines, and F1 progeny of the CBP-expressing transgenic rice lines crossed with AVP1-expressing transgenic rice lines for drought tolerance.
- Test AtAVP1, and maize CBP (calcium binding domain of careticulin) transgenic rice lines, and F1 progeny of the CBP-expressing transgenic rice lines crossed with AVP1-expressing transgenic rice lines for salt stress tolerance.
Virginia State University - Shuxin Ren
Objective 1: Test the null hypothesis that there is no difference in field growth characteristics and seed traits (maturity, height, lodging resistance, seed yield, protein concentration, amino acid composition of the protein, oil concentration, fatty acid composition of the oil, and inorganic phosphorous content) between TN09-239 and USG 5601T.
Objective 2: Test the hypothesis that DNA collected from vegetative leaf tissue can distinguish whether the plant will produce seeds at harvest that will contain normal versus low levels of phyate, by evaluating two genetic loci: SSR markers SATT237 (chromosome 3, linkage group N) and SATT561 (chromosome 19, linkage group L).
Objective 3: Extend the knowledge of SSR molecular markers to develop a test of single nucleotide polymorphisms (SNPs) at two genetic loci controlling soybean seed phytate.
North Carolina A&T State University - Guochen Yang
Objectives:
- To perform salt tolerance assays of transgenic rice plants in collaboration with NCSU's CPBR-funded project.
- Student training through fellowship: actively engaged students in the area of plant tissue culture related to transformation, molecular breeding and environmental research.
University of Tennessee - Zong-Ming Cheng
Objective 1: To characterize the transgenic plants for enhanced tolerance to high and low temperature, drought and flooding.
Objective 2: To determine whether the PtXTH22 promoter can be used as a broad stress-inducible promoter.
Objective 3: To characterize the promoter of the PtXTH22 gene.
Iowa State University - Michael Kessler
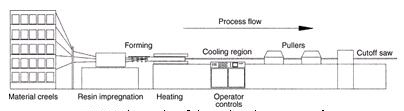
The goal of this project is to develop bio-based resins, prepared by co-polymerization of agricultural oils (e.g., corn, linseed, and soybean) and derivatives, with commercially available co-monomers, for the manufacture of fiberglass-reinforced composites used in pultrusion processing. Pultrusion is a continuous manufacturing process for creating composites with a constant cross-section. In the pultrusion process (shown schematically in Figure 1 below), the fiber reinforcement is impregnated with the resin matrix and pulled through a heated die where it is shaped and cured in one continuous step.
Figue 1.Schematic of the pultrusion process
The specific objectives for the 2-year project are the following:
- develop appropriate resin formulations with the right combination of processing viscosity, cure kinetics, and ultimate thermo-mechanical properties for pultrusion processing.
- determine the best processing conditions for the pultruded fiberglass/bio-resin systems in order to maximize the thermo-mechanical properties of the composites and economics of the system, and
- evaluate the long-term strength and durability of the composites compared to existing polyester materials through accelerated aging and environmental exposure testing.
University of Georgia - Andrew Paterson
Objectives:
- Paired-end sequencing of about 120,000 BACs, roughly equally sampling the two libraries described above, to be done by subcontract to a fee-for-service vendor selected on a competitive basis.
- Computational analysis of the resulting sequence assembly by several tests to identify possible errors, conducted in the PI's lab as described (Paterson, et al., 2009), including:
- Comparison to the order of sequence-tagged-sites along the (~1 cM density) cotton genetic map (Rong, et al., 2004), to investigate accuracy across regions of several cM or more;
- Comparison to the order of overgo and BAC-end sequences in BAC clones comprising the physical map (detailed above), testing contiguity of regions of less than the ~1 cM resolution of the genetic map, down to the ~100 kb average size of a BAC;
- Comparison to the Arabidopsis genome, the most complete angiosperm genome and the closest relative of cotton that has been fully sequenced. We have previously shown appreciable microsynteny and even some regions of macrosynteny between Arabidopsis and cotton (Rong, et al., 2005a). A sequence scaffold would not be expected to show long stretches of cotton-Arabidopsis synteny if incorrectly assembled. However, because there will surely be many (hundreds?) small rearrangements between these taxa, synteny information will be used only to support other lines of evidence, and the assembly will never exclusively rely on synteny to support any genome arrangement.
Georgia Institute of Technology - Gerald Pullman
Hypothesis 1: Natural redox agents control redox potential in developing female gametophyte and embryo.
Objective 1: Use LC/MS and LC/MS/MS to confirm and extend preliminary results to identify and profile ascorbic acid (reduced), dehydroascorbate (oxidized), glutathione (reduced) and glutathione disulfide (oxidized) in female gametophyte and embryo tissues.
Hypothesis 2: Redox potential controls somatic embryo development in vitro.
Objective 2: Run early-stage, maturation, and germination somatic embryo growth tests with natural redox chemicals.
Objective 3: Run early-stage, maturation, and germination somatic embryo growth tests with alternative redox chemicals.
Objective 4: Measure ascorbic acid (reduced), dehydroascorbate (oxidized), glutathione (reduced) and glutathione disulfide (oxidized) in somatic embryos to determine if target concentrations found in zygotic embryos are achieved during growth in vitro.
Hypothesis 3: Medium redox potential changes over time due to air exposure, tissue growth and medium components.
Objective 5: Determine medium redox potential change over time due to air exposure and tissue growth.
Objective 6: Develop methods to control medium redox potential to desired levels over time and in presence of growing somatic embryos.
Progress Summary:
Iowa State University- Bryony C. Bonning
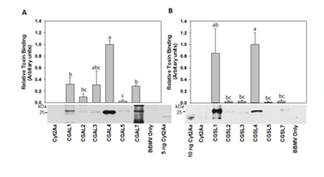
Figure 1. CGALn and CGSLn bind to pea aphid gut BBMV
more strongly than Cyt2aAa. (A) Pull down assays were
conducted following incubation of activated Cyt2Aa, CGALn
or CGSLn and pea phid gut BBMV. Membrane bound toxin
wsa detected by western blot with anti-Cyt2Aa antiserum.
Western blot images (below) were scanned and processed
using ImageJ to estimate the relative amount of activated
toxin in associated with pea aphid BBMV. Relative toxin
binding is shown for the mean ± SE for two pull-down
assays. DIfferent letters indicate the statistical significance
at p ≤ 0.05 (one-way ANOVA).
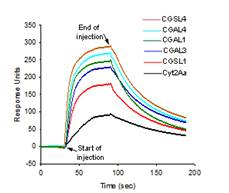
Figure 2. BIAcore surface plasmon analysis
of toxin binding to small unilamellarvesicles (SUV).
Sensorgram showing the real-time interaction
between 6 µm Cyt2Aa, CGAL1, CGAL3, CGAL4,
CGSL1, CGSL4 and immobilized pea aphid gut
membrane SUV. L1 chip surfaces were prepared
with 4999 RU of ligands. The sata shown are
representative of two independent experiments.
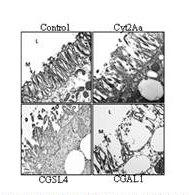
Figure 3. CGAL1 and CGSL4 cause
extensive damate to the pea aphid
gut epithelium. Transmission electron
micrographs show the intact apical surface
of the gut epithelial membrane with
microvilli (NM) projecting into the gut lumen
(L) in aphids fed on control diet (Control).
The microvilli of Cyt2As-fed asphids showed
some damage, consistent with the low level of
toxicity seen again A.pisum with this toxin. The
integrity of the gut phithelia of aphids fed on
CGAL1 and CGSL4 was severely compromised.
Mocrographs were taken at the same
magnification with the 1 µm scale indicated
for the Control.

Figure 4. Relative binding of wild type
Cyt2Aa, CGAL134 and CGSL14 to pea aphid
gut BBMV in pull down assays.
| Genotype | Mat | yld bu/A | tld rank | LG N Chomosome 2 melting temperature | LG L Chromosome 19 melting temperature | µg Pig-1 | Growth habit | Protein |
|---|---|---|---|---|---|---|---|---|
| 5601T | 282 | 69.2 | 1 | 57 0C | 60 0C | 201 | dd | 40.7 |
| TB09-239 | 281 | 62.4 | 7 | 62 0C | 54 0C | 1560 | DD | 41.8 |
| 56CX-1273 | 280 | 67.0 | 2 | 62 0C | 54 0C | 1808 | dd | 41.3 |
| 56CX-1283 | 281 | 66.8 | 3 | 62 0C | 54 0C | 1555 | dd | 41.9 |
Table 1. Effective genotyping helped us select these low phytate lines with improved
agronomic characteristic
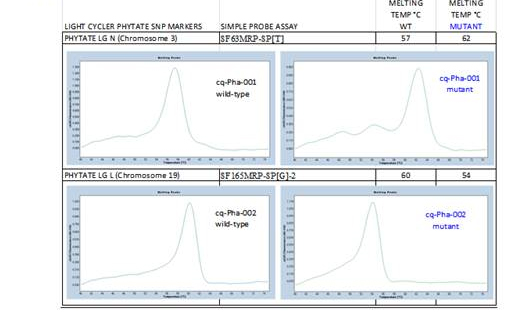
Figure 1. SNP Melting Curves
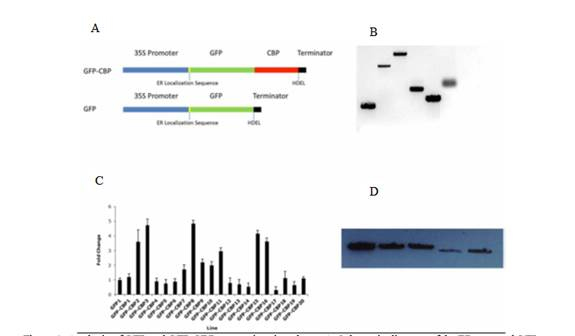
Figure 1. Analysis of FGP and FGP-CCP-expressing rice plants. A. Schematic diagram of the
ER-targed GFP-CBP and its control, fgP. B. Southern blot analysis of six independent transgenic lines.
They are )left to right): FPB-CBP 8, 4, 7. 12, 14 and FGP-Lowl1, a FFP transgenetic line as a control
(DIG-labled probe: 700 bp FGP coding sequence). C. qRT-PCR analysis to determine GFP or FGP-CBP
expression in 20 independent transgenetic rice plants. D. Western blot analysis of five GFP-CBP
transgenic lines. They are FGP-CBP 8, 15, 16,04 and 7. Anitbody against FGP was used for the Western
blot. No breakdown products were apparent
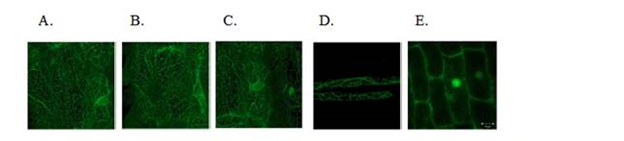
Figure 2. The GFP-CBP fusion protein localization in the endoplasmatic reticulum was verified
using a Zeiss confocal laser scanning microscope. A - C, rice root cells in ER-GFP only (A) and
ER-FGP-CBP (B and C) transgenetic rice seedlings, D, root hairs of a GFP-CBP transgenetic rice
seedling, and E; rice root cells of non-ER-targeted GFP transgenetic plants. Rice seeds were
germinated on half strength MS media and grown for one week. 2 cm rice roots were taken from
the rice seedlings and mounted on a microscope slide.

Figure 3. TOtal calcium content in control rice plants and AVP1-, GFP-, CGP-CBP- and GFPCBPx AVP1
transgenetic rice plant lines. A. Total calcium content in FGP-CBP transgenetic rice plants is increased.
Total calcium content in the rice leaves of each transgenetic plant lines was measured three times from three
different biological samples. * means they are significanly different (P<0.05). B. Control and transgenetic
rice plants exhibiting normal phenotypes 6 weeks after germination. From left to right: 1. wild type, 2: GFP,
3. AVP1-1, 4. AVP1-2, 5: GFP-CBP Low, 6: GFP-CPB High, 7: GFP-CBPlow % AVP1-2, 9: FGPCBP High %
AVP1-1, 10: GFP-CBP High % AVP1-2
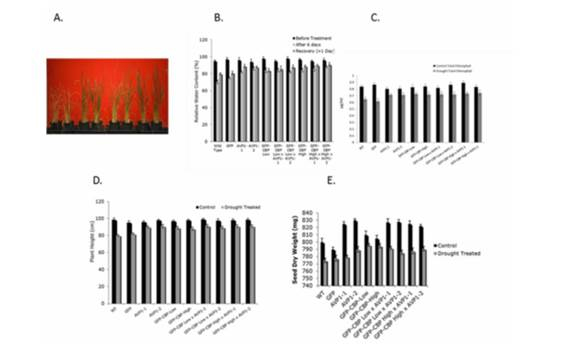
Figure 4. A: 1, WT, 2, GFP,3. AVP1-1, 4. AVP1-2, 5. GFP-CBP-Low, 6. GFP-CBP-High, 7.
GFP-CBO-Low % AVP1-1, 8: GFP-CBP-High % AVP1-2, Plants were not watered for 6 days
and then watered. The picture taken wafer withholding water for another 6 days after watering
them. B: Relative water content of transgenic and control rice plants. They were measured three
times; one time point before intermittent drought treatment and two time pints during intermittent
water stress. C. Chlorophys content of transgenic and control rice plants. The chlorophyl content
was measured before and after drought treatments. D. The longest shoot length of control and
drought treated rice plants. E. The seed weight of seeds per panide in 10 different transgenic rice
lines. All of the transgenic lines show higher seed weight compared to the control lnes.
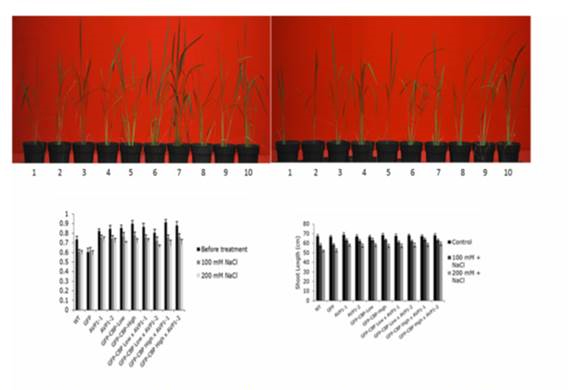
Figure 5. AVP1 and GFP-CBP shows increased salt tolerance. A and B, After 12 days of
100 mM(Fig a) and 200 mM (Fig b) NaCl treatment. 1. Wt, 2. GFP, 3. AVP1-1, 4. AVP1-2, 5.
GFP-CBP-Low, 6. GFP-CBP-High, 7. GFP-CBP-Low % AVP1-1, 8. GFP-CBP-Low % AVP1-2, 9.
GFP-CBP-High % AVP1-1, 10. FGP-CBP-High % AVP1-2. C, chlorophyl content of transgenic
and control rice plants. The chlorophyl content was measured at two different concentrations of
NaCi before and after a period of 12 days. D, Shoot height of control and salttreated rice plants.
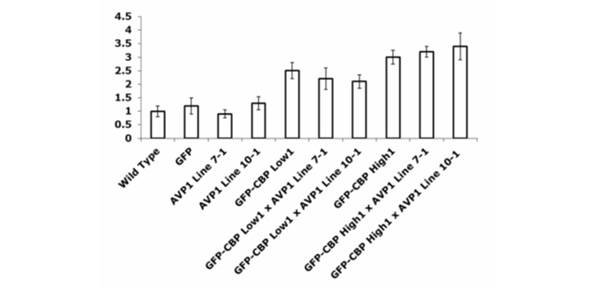
Figure 6. Rice CIPK5 (Os)1g0206700), a homologue of Arabidopsis AtCIPK6, is
unpregulated in transgenic plants expressing CBP, indicating a role in CBP-mediated
drought/salt tolerance. The left axis shows fold change relateive to wild type rice.
Error bars represent the standard error of three different experiments.
This project was implemented at the Virginia State University from mid of 2010 to the end of 2012. Soybean lines, TN09-239 and USG5601T, were provided by Dr. Pantalone at University Tennessee and grown in VSU Randolph Farm in 2011 and 2012 in a randomized complete block design, with four row plots. Agronomic traits were recorded and compared between TN09-239 and USG5601T.
Both lines showed white flower and gray pubescence. In addition, they all have same flowering date and are resistant to shattering (score 1). TN09-239 and USG 5601T are all in maturity group 5 (MG V), however TN09-239 is in early MG V, while USG 5601T is in late MG V. In terms of 100 seeds weight, TN09-239 is little bit higher (22 g) than 5601T (21 g) but no significant difference. Although these two soybean lines were considered as near isogenic line with only difference at seedphytate level, we do find that the plant heights between TN09-239 and 5601T were significantly different with TN09-239 of 131.6 cm and 5601T of 68.0 cm. Because of this height difference, TN09-239 appeared more susceptible to lodging stress. This plant height difference may due to environmental difference. However, under this experimental design, we only include one location. Further evaluation at multi-locations on this issue is required to confirm this result. The yields of these two soybean lines showed no significant difference.
In addition to agronomic traits evaluated in the field, the harvested soybean seeds also were analyzed for protein concentration, oil concentration, fatty acid composition of the oil, and seed phosphorous contents. Total protein content in TN-239 seed is 42.06% on average, and in 5601T seed is 41.36% on average. No significant difference was observed. Total seeds oil content for TN239 is 18.45%, and 5601T is 18.59%. Again, no significant difference was observed. Oil fatty acid composition results showed that TN-239 contains significant more 18:0 fatty acid (4.0%) than 5601T (3.6%). On the other hand, TN-239 has significantly less (6.95%) 18:3 fatty acids than that of 5601T (7.87%). All other oil fatty acid compositions showed no difference between these lines.
Total P content in seeds also was evaluated in two biologically duplicated samples. Results showed that TN-239 has significantly higher phosphate level (1.31g/kg seed) that 5601T (0.52g/kg seed). However, the phytate contents in the seeds are variable from repeat to repeat with the trend that TN-239 has less phytate than 5601T. More investigation is needed to make a solid conclusion on seed phytate content.
We also collected leaves from all plots and isolated DNA from each collected sample. SSR markers SATT237 and SATT561 were used to determine the phytate levels between TN09-239 and USG 5601T. Although the PCR products were successfully amplified from both varieties, separation on 4% agrose gel could not distinguish these two varieties. SNP markers were used to identify genotypes that control phytate levels. As expected, SNP markers used can distinguish these two lines at loci controlling phytate levels.
On the educational propose of this fellowship, the student, together with PIs were traveled to TN. The student was successfully trained on DNA technology, including different DNA isolation methods, SSR markers and SNP markers. Exposure to these state-of-the-art biotechnology facilities will give the student more understanding of the application of biotechnology to agriculture production and stimulate the student’s passion to pursue a biotechnology-related education and career. In fact, the first student who was recruitied has now graduated and has secured funding to continue her education at the graduate level. In addition, a second undergraduate student who was recruited to the project, will be well trained on soybean breeding related techniques upon completion of this project.
North Carolina A&T State University
University of Tennessee
The work in this year has been focused on exploring the mechanism of stress tolerance in transgenic plants over-expressing PtXTH22 gene and functional characterization of the PtXTH22 gene promoter.
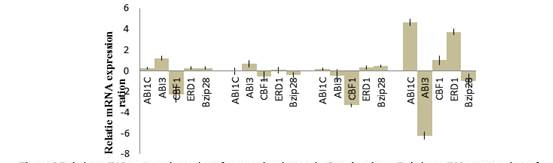
Figure 5. Relative mRNA expression ratios of stress-related genes in Popuhus plants. Relative
mRNA expression opf the gene was represresented as the value in the transgenic lines relative to
value in the wild type. Line X was ABA-treated or untreated line 18 or 2-B, WT was represented by
wild type, and the target was the ABI1C, ABI3, CBF1, ERD1, bZIP28 genes. The value in Y-axis is -ΔΔCt.
The whole plants were collected from 4-week subcultured plants.
The progress made toward each objective is described below:
- A series of bio-based resins have been investigated. Ring-opening metathesis polymerization (ROMP)-based copolymer of Dilulin (a modified linseed oil) and dicyclopentadiene (DCPD) and cationic-based copolymer of soybean oil, styrene and divinylbenzene have been selected as potential candidates for the pultrusion process. Resin formulations of these two systems were optimized to meet special requirements of pultrusion fabrication method, including rapid polymerization rate and thermomechanical properties of the cured resin. ROMP-based bioresins show promising high toughness, with a 3-fold improvement in toughness over the benchmark petroleum-based unsaturated polyester resin. Additional characterizations have been carried out to study the fracture toughness mechanism of the ROMP-based bioresin. To obtain best properties of fiberglass/ROMP-based bioresin composites, several silane coupling agents were applied to modify the glass fiber surface. Thermal and mechanical characterizations show that interfacial strength of the composite was improved significantly with norbornenylethyldimethylchlorosilane (MCS). This silane will also be applied in the pultrusion process to improve the properties of pultruded composites
- A table-top pultrusion system has been set up in our lab.We have successfully made prototype fiberglass samples with both commercial polyester and Dilulin/DCPD bioresin. Improvement of the pultrusion system to obtain high quality pultruded samples with a smooth outer surface is still in progress.
- Long term performance evaluations of ROMP-based Dilulin/DCPDresin and glass fiber composite have been carried out with QUV and XENON accelerated weathering test chambers.
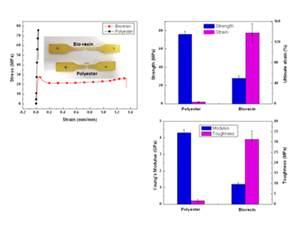
Figure 2. (left) Representative e stress-strain curves for bio-based resin
and unsaturated polyester resin. (right) while the strength and stiffless
toughness is significanly higher for hte bio-based resin.
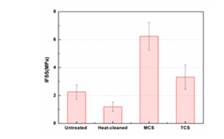
FIgure 3. Interfacial shear strength (IFSS) for different types of fibers

Figure 4. Change is polymer appearnce afgter expsure t
from 0 (left) to 100h (right)

Figure 5. Bench topo pultrusion syustem deisgned and built for this
project for continous manufacturing of bio-resin fiberglass composites.

Figure 6. Polyester and bio-resin samples made by the pultrusion process.
Future Activities:
University of Tennessee
Now that we have the determinate linkage resolved, we can increase seed and follow up on testing for yield and nutrition for the trait.
North Carolina State University
The project is completed.
North Carolina A&T State University
Analyzing data, and preparing abstracts and manuscripts for presentations and/or publications.
University of Tennessee
- Determinethe mechanism for drought and ABA tolerance of transgenic plants.
- Test the transgenic soybean for drought tolerance.
- Further test the pXTH22:GUS transgenic Arabidopsis to determine whether the promoter is multi-stress inducible.
- Test different sections of promoter to determine which sections of promoter is responding different stresses.
Iowa State University
In the upcoming year, we plan to (1) continue to improve the process ability and appearance quality of pultruded fiberglass/Dil30DCPD70 composites with our table-top pultrusion machine; (2) evaluate short and long term strength and durability of the pultruded fiberglass/Dil30DCPD70 composites; (3) apply silane coupling agent in fiberglass/Dil30DCPD70 system to further improve the properties of the pultruded composites; and (4) apply the cationic polymerization resin system in thepultrusion process.
More specifically, we have carried out many trials with the ROMP-based Dil30DCPD70 bioresin system on our table-top pultrusion machine. The resulting pultruded composites have a rough surface, effecting the appearance of the composite material. This surface roughness is believed to be caused by a stick-slip progression during processing. Our plans for the coming year are to continue optimizing the processing parameters and to find the best combination of pulling rate, cure temperature and fiber ratio to produce pultruded composites with perfect profile shape and smooth surface. At the same time, we also will add some release agents into the resin. There are a wide range of commercially available release agents in the market. Finding the most suitable type and additive amount of the release agent, as well as the effect of release agents on final pultruded products also will be a part of our future work.
Once we can get good samples from repeatable trials, we will start to characterize the properties of the pultruded composites and compare them to the existing unsaturated polyester pultruded materials. A series of engineering properties that are necessary for structural materials will be evaluated following ASTM standards, such as tensile strength, Young’s modulus, flexure strength, flexure modulus, coefficient of thermal expansion, impact behavior, and compressive properties. The long-term performance of the bio-based pultruded composites will be evaluated with the help of our industrial partner and their extensive environmental chemistry and product test laboratories. Materials will be subjected to a range of elevated temperature and environmental conditions, including humidity, ultraviolet radiation, and full-spectrum sunlight. The effects of those conditions on discoloration, swelling, warping, delaminating, and change in mechanical properties will be analyzed systematically. Because we already studied the long-term performance of the neat resin in the first year of the study, these results will be helpful to gain an understanding of the effect of reinforcement on the long-term performance of pultruded composites.
In the first year of the project, we have investigated the effect of using silane coupling agents on improving the compatibility and adhesive strength between the glass fibers and the bio-based resin. Our studies indicate that interfacial strength between glass fiber and Dil30DCPD70 bio-matrix is increased significantly with the application of norbornenylethyldimethylchlorosilane coupling agent. So applying this silane in the pultrusion process will further enhance properties of the final pultruded composites. The challenge is developing a process for applying the silane in a continuous pultrurion process. Usually, in industry, silane coupling agent can be applied in a solution or in vapor phase. We will compare these two methods first in batch processed conditions that involve treating the glass fiber with silane solution or silane vapor prior to the pultrusion process. Then we will try to add the silane coupling agent during the pultrusion process. Special equipment will be designed to fit the whole continuous pultrusion process. Taking the silane solution method for example, a silane solution bath and a drying tube furnace will be implemented before resin bath in the pultrusion process line.
Our efforts in the second year also will be focused on the cationic polymerization resin system - soybean oil, styrene and divinylbenzeneapplied to the pultrusion process. Following similar experiments as used for the ROMP-based bioresin, processing parameters and a suitable release agent will be optimized first. One challenge for cationic-based bioresin is stabilization of the resin after mixing with initiator. It is difficult for this resin to remain stable at room temperature after it has been mixed with the cationic initiator, especially in large amounts where cure exothermic conditions can increase the temperature of the resin bath. Lowering the resin storage temperature is one approach to increasing pot life of the resin. Towards this end, a special resin bath will be designed for the cationic resin system. Once we can get good samples from pultrution, short-term and long-term performance will be evaluated and also compared with unsaturated polyester materials and the ROMP resin system.
University of Georgia
Paired-end sequencing of about 120,000 BACs – this has been completed. Indeed, while our proposal was under consideration by CPBR, another (public) entity supported end-sequencing of about 60,000 BACs from the G. raimondii HindIII library. Accordingly, the PI and representatives of the matching company (Bayer) agreed to an accordingly reduced investment in BAC end sequencing (that has been met through a contract with a third party, the Hudson Alpha Institute) … redirecting the remaining resources to reduced-representation resequencing of about 30 cotton genotypes, about 14 of which are selected by Bayer for proprietary reasons. The remainder will be selected from publicly available germplasm by the PI but in consideration of feedback from Bayer, giving preference to genotypes from which there exist other publicly available resources (for example Pima S6 and Acala Maxxa have BAC libraries, and G. barbadense K101 was used in reference genetic mapping), and/or which are parents of populations that are being made toward a cotton nested association mapping (NAM) resource. Several of the public lines have been tentatively completed, although data validation remains pending. Bayer is preparing DNA of its lines for shipment to the PI.
Computational analysis of the resulting sequence assembly by several tests to identify possible errors, conducted in the PI’s lab as described (Paterson, et al., 2009), including:
- Comparison to the order of sequence-tagged-sites along the (~1 cM density) cotton genetic map (Rong, et al., 2004), to investigate accuracy across regions of several cM or more.
- Comparison to the order of overgo and BAC-end sequences in BAC clones comprising the physical map (detailed above), testing contiguity of regions of less than the ~1 cM resolution of the genetic map, down to the ~100 kb average size of a BAC,
- Comparison to the Arabidopsis genome, the most complete angiosperm genome and the closest relative of cotton that has been fully sequenced. We have previously shown appreciable microsynteny and even some regions of macrosynteny between Arabidopsis and cotton (Rong, et al., 2005a). A sequence scaffold would not be expected to show long stretches of cotton-Arabidopsis synteny if incorrectly assembled. However, because there will surely be many (hundreds?) small rearrangements between these taxa, synteny information will be used only to support other lines of evidence, and the assembly will never exclusively rely on synteny to support any genome arrangement.
Items a and b, above, were completed in advance of public release (under Ft Lauderdale principles) of the genome assembly in January 2012.
Item c was completed as an element of the primary description of the cotton reference genome sequence (Paterson, et al., 2012), confirming early results supportive of the high quality of assembly, and revealing an unexpected dimension of cotton genome evolution, specifically that it has experienced a paleopolyploidy of complexity previously unprecedented … it remains unclear whether this resulted in an aggregate 5x or 6x increase of cotton chromosome numbers, and whether it was a single event or two very closely spaced events (for example 3x followed by 2x).
References:
Febvay, G., B. Delobel & Y. Rahbe (1988) Influence of the amino-acid balance on the improvement of an artificial diet for a biotype of Acyrthosiphon-pisum (Homoptera, Aphididae). Canadian Journal of Zoology-Revue Canadienne De Zoologie, 66, 2449-2453.
Li, J., Koni, P. A. and Ellar, D. J. (1996). Structure of the mosquitocidal delta-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J. Mol. Biol. 257, 129-152.
Liu, S., Sivakumar, S., Sparks, W.O., Miller, W.A., Bonning, B.C. (2010). A peptide that binds the pea aphid gut impedes entry of Pea enation mosaic virus into the aphid hemocoel. Virology 401 (1): 107-16.
Pérez C., Fernandez L.E., Sun J., Folch J.L., Gill S.S., Soberón M. (2005). Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. PNAS, vol. 102 no. 51 18303-18308.
Journal Articles: 10 Displayed | Download in RIS Format
| Other center views: | All 32 publications | 11 publications in selected types | All 10 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Chougule NP, Li H, Liu S, Linz LB, Narva KE, Meade T, Bonning BC. Retargeting of the Bacillus thuringiensis toxin Cyt2Aa against hemipteran insect pests. Proceedings of the National Academy of Sciences of the United States of America 2013;110(21):8465-8470. |
EM834388 (2012) |
Exit Exit Exit |
|
|
Cui H, Kessler MR. Glass fiber reinforced ROMP-based bio-renewable polymers: enhancement of the interface with silane coupling agents. Composites Science and Technology 2012;72(11):1264-1272. |
EM834388 (2012) EM834388 (Final) |
Exit Exit |
|
|
Madbouly SA, Xia Y, Kessler MR. Rheokinetics of ring-opening metathesis polymerization of bio-based castor oil thermoset. Macromolecules 2012;45(19):7729-7739. |
EM834388 (2012) |
Exit |
|
|
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012;492(7429):423-427. |
EM834388 (2012) |
Exit Exit Exit |
|
|
Thunga M, Xia Y, Gohs U, Heinrich G, Larock RC, Kessler MR. Influence of electron beam irradiation on the mechanical properties of vegetable-oil-based biopolymers. Macromolecular Materials and Engineering 2012;297(8):799-806. |
EM834388 (2012) EM834388 (Final) |
Exit |
|
|
Ye X, Yuan S, Guo H, Chen F, Tuskan GA, Cheng Z-M. Evolution and divergence in the coding and promoter regions of the Populus gene family encoding xyloglucan endotransglycosylase/hydrolases. Tree Genetics & Genomes 2012;8(1):177-194. |
EM834388 (2012) EM834388 (Final) |
Exit Exit Exit |
|
|
Ye X, Busov V, Zhao N, Meilan R, McDonnell LM, Coleman HD, Mansfield SD, Chen F, Li Y, Cheng ZM. Transgenic Populus trees for forest products, bioenergy, and functional genomics. Critical Reviews in Plant Sciences 2011;30(5):415-434. |
EM834388 (Final) |
Exit |
|
|
Cui H, Hanus R, Kessler MR. Degradation of ROMP-based bio-renewable polymers by UV radiation. Polymer Degradation and Stability 2013;98(11):2357-2365. |
EM834388 (Final) |
Exit |
|
|
Lin W, Hagen E, Fulcher A, Hren MT, Cheng ZM. Overexpressing the ZmDof1 gene in Populus does not improve growth and nitrogen assimilation under low-nitrogen conditions. Plant Cell, Tissue and Organ Culture (PCTOC) 2013;113:51-61. |
EM834388 (Final) |
Exit |
|
|
Lin WL, Cai B, Cheng ZM. Identification and characterization of lineage-specific genes in Populus trichocarpa. Plant Cell, Tissue and Organ Culture (PCTOC) 2014;116:217-225. |
EM834388 (Final) |
Exit |
Supplemental Keywords:
Bt toxin; aphid control, phytic acid, phosphorous nutrient load, eutrophication, clean water, calcium, chlorophyll, CIPK, RWC, salt, marker-assisted selection; low phytate soybean; molecular breeding, agronomic traits, chlorophyll, electrolyte leakage, proline, protein, relative water content, stress tolerance, sustainability, bio-based, polymers, composites, agricultural oils, pultrusion, somatic embryogenesis, redox chemicals, oxidation, reductionRelevant Websites:
Progress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.