Grantee Research Project Results
2010 Progress Report: The Texas-Indiana Virtual STAR Center; Data-Generating in vitro and in silico Models of Developmental Toxicity in Embryonic Stem Cells and Zebrafish
EPA Grant Number: R834289Center: Organotypic Culture Models For Predictive Toxicology Center
Center Director: Rusyn, Ivan
Title: The Texas-Indiana Virtual STAR Center; Data-Generating in vitro and in silico Models of Developmental Toxicity in Embryonic Stem Cells and Zebrafish
Investigators: Gustafsson , Jan-Ake , Finnell, Richard H. , Glazier, James A
Institution: University of Houston - University Park , Indiana University - Bloomington , Texas A & M University
EPA Project Officer: Hahn, Intaek
Project Period: November 1, 2009 through October 31, 2012
Project Period Covered by this Report: November 1, 2009 through October 31,2010
Project Amount: $3,190,993
RFA: Computational Toxicology Research Centers: in vitro and in silico Models Of Developmental Toxicity Pathways (2009) RFA Text | Recipients Lists
Research Category: Computational Toxicology , Human Health , Safer Chemicals
Objective:
- Generate developmental models suitable for high throughput screening.
- Generate high information content models on development and differentiation.
- Develop computational models for developmental toxicology with the ultimate aims of first recreating normal development (in wild-type) and then classifying possible mechanisms by which chemical perturbations cause experimentally observed developmental defects.
- Perform proof-of-concept experiments of the in vitro and in silico test platforms with a blind test of chemicals.
- Work towards a global harmonization of chemical regulation.
A. Collaborate with OECD regarding validation and adoption of new methods for the OECD guidelines.
B. Transfer of TIVS Center results for mathematical calculations of dose-effects regulators in the USA and Europe.
C. Transfer of TIVS results to the public databases.
D. Disseminate results on chemical screening to stakeholders and the general public.
- Education of a new generation of multidisciplinary scientists in the fields of chemical screening, computational toxicology and risk assessment
A. Three workshops (one/year)
B. Co-supervision of students/postdocs
- Efficient communication with and timely delivery of reports to the US-EPA/NCER.
- Efficient collaboration with other parties in the field of chemical screening.
- Continuous information to the TIVS Center partners about project progress and results, as well as quality assurance activities.
- Provision of assistance and advice to the TIVS Center partners regarding administrative issues and reporting routines.
- Identification of potential obstacles and provision of appropriate solutions.
- Review and evaluation of the research performed within the TIVS Center.
- Efficient fiscal management of the subcontracts to research sites.
- Advertise, interview and hire postdocs and lab technician.
- Collect zebrafish screening models already present in the zebrafish research community.
- Produce new transgenic zebrafish lines.
- Imaging the normal development of vasculature, somites and axon pathfinding in 2D and 3D.
- Exchange of data with Partners 2 and 3 to generate in silico models.
- Generate automation and imaging strategy for toxicity screens using a panel of known chemicals and transgenic fish.
- Treatment with test chemicals.
- Advertise, interview and hire postdocs and lab technician.
- Expand ES cell clones from TIGM library.
- Optimize models of ES cell differentiation.
- Capture images using newly acquired In Cell technology.
- Treatment with test chemicals.
Progress Summary:
- Start up meeting November 19-20, 2009 at UH.
- Computational Toxicology Subcommittee, September 29-30, 2009. Attended by Maria Bondesson, Richard Finnell and James Glazier. Poster Presentation (EPA).
- Computational Toxicology Centers STAR Progress Review Workshop, October 1, 2009, EPA Attended by Maria Bondesson, Richard Finnell and James Glazier. Oral Presentation (EPA).
- Screening models meeting at EPA, May 18, 2010.
- Regular meetings between UH and Texas A&M/University of Texas at Austin.
- Regular telephone meetings every second week between UH and IU.
- February 26, 2010 with all participants, EPA, QMP Manager
- November 1, 2010 with all participants and EPA
- September 22, 2010 with UH, IU and research group of Fatima Merchant to discuss imaging of vascular development in zebrafish
- October 27, 2010 with UH, IU and research group of Fatima Merchant.
- Maria Bondesson is member of Advisory Board of ChemScreen.
- Bart van der Burg is member of Advisory Board of TIVS Center.
- Maria Bondesson participated in ChemScreen start up meeting February 1st, 2010, Amsterdam, The Netherlands.
- Maria Bondesson participated in ChemScreen meeting September 15-16, 2010, Zeist, The Netherlands.
- Meeting with CASCADE Acert (Ingemar Pongratz) Houston, December 2, 2010.
- Netherlands Bioinformatics Center, Workshop Modeling Angiogenesis: Joining Cells, Maths and Computers Oct 4-8, 2010 Leiden, The Netherlands (Swat, Glazier).
- Jan-Ake Gustafsson spoke at the SAFE Consortium Workshop: ‘Dietary Exposure to Endocrine-Active Pesticides' Brussels, Belgium. 26-27 November 2009.
- Maria Bondesson attended the Crescendo Integrated Project annual meeting May 27-29, 2010, in Munich, Germany, on Nuclear receptors during development and aging.
| # | Gene | Used for | Reporter | Start time of expected expressioin (hpf) | Status |
|---|---|---|---|---|---|
| Transgenic fish in proposal | |||||
| 1 | goosecoid | Early patterning, epiboly, early cell movements and developmental delay. | GFP- | 3.5 hpf | Tg[gsc:Gal4-VP16]/Tg[USA-GFP] in zf community. Targeted gene expression in the zebrafish percholdal plate. Inbal A, Topczewski J, Solnica-Krezel L. Genesis. 2006 44(12): 854-8. |
| 2 | gharma | Early patterning, epiboly, early cell movements and developmental delay. | EGFP | 3.5 hpf | With us (made in house) |
| 3 | bmp2b | Patterning (anterior-posterior symmetry), early cell movements. | GFP- | 1 cell state 0 hpf (maternal contributed) | Not done |
| 4 | wnt8 | Patterning (anterior-posterior symmetry), early cell movements | GFP- | 1 cell state 0 hpf (maternal contributed) | not done |
| 5 | bmp4 | Patterning (left-right symmetry) | GFP- | 10 hpf | Present in zf community. Proximal upstream region of zebrafish bone morphogenetic protein 4 promoter directs heart expression of green flourescent protein. Shentu H, Wen HJ, Her GM, Huang CJ, Wu JL, Hwang SP. Genesis. 2003 37(3):103-12 |
| 6 | ng1 | Neurogenisis, axon guidance, early developmental delay | GFP | 10 hpf | With us |
| 7 | fli1 | Angiongenesis and blood vessel remodeling, hart morphology and function. | EGFP- | 11 hpf | With us |
| 8 | flk1 | Angiogenesis and blood vessel remodeling, heart morphology and function. Expressed in tip cells. | EGFP- | 11 hpf | With us |
| 9 | Unc5b | Blood vessel formation, expressed in tip cells at the forefront of arterial and venous sprouts. | RFP- | 9 hpf | Not necessary; Tip vessel cells are seen in the Flk-1 fish. |
| 10 | unc45bi | A myosin chaperone, Muscle develpment and somitogenesis. | GFP | 9 hpf | With us |
| Additional transgenic fish | |||||
| 11 | Gata-1 | Red blood cells | dsRed | With us | |
| 12 | Smych-1 | Slow myosin heavy chain 1 Muscle developmnt and somitogenesis | GFP | With us | |
| 13 | HGn39B | Muscle expression Muscle development and somitogenesis | GFP | With us | |
| 14 | HGn50D | Yold expression Yold utilization | GFP | With us | |
| 15 | ERE | Estrogen response element driven reporter | GFP | With us | |
| Mutant fish | |||||
| 16 | Casper | Transparent | With us | ||
| 17 | Nacre | Transparent | With us | ||
| 18 | Albino | Transparent | With us | ||
| WT strains | |||||
| 19 | DZ | With us | |||
| 20 | WIK | With us | |||
| 21 | AB | With us | |||
| 22 | Tuebingen AB | With us | |||
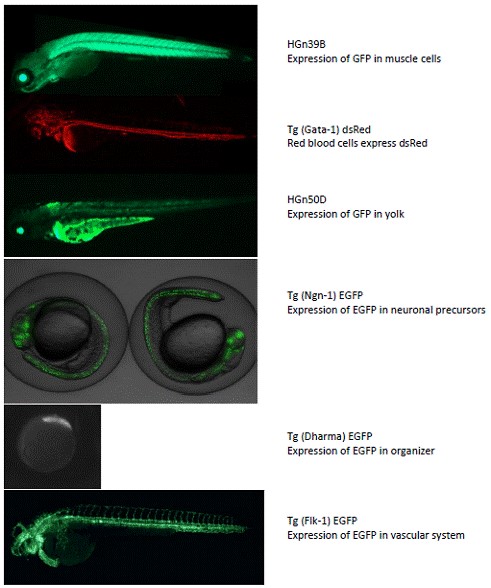
Figure 2. Fluorescent microscopy of selected transgenic fish present in the
zebrafish faciluty at UH.
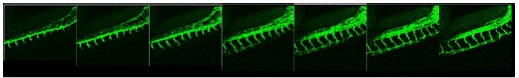
Figure 2. Development of intersegmental vessels in the zebrafish trunk from 26 to 36 hours
post fertilization (hpf) in the Tg (Flk-1) EGFP fish. Lateral view, dorsal side is down and
anterior to the left.
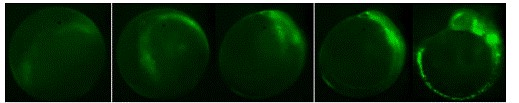
Figure 3. Development of Ngn-1 EFGP transgenic fish showing EGFP expression in
neuronal precurors between approximately the 1 somite state through 10 somite stage.
Lateral view, anterior up.
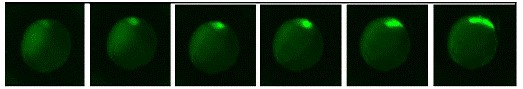
Figure 4. Recording of dharma deiven EGFP expression in developing Dharma EGFP
transgenic fish between approximately the sphere state thorugh 80% epiboly.
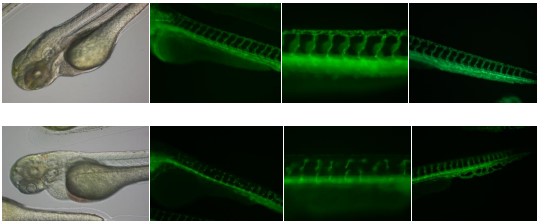
Figure 5. Zebrafish embryos exposed to 3nM sodium arsenite from 1 hpf to 72 hpf. Picutres
taken on Olympus IX51 microscope.
| As (1mg/mL) exposure time | 0-72 hpf** | 24-48 hpf | 48-72 hpf |
| Pericardial exdema | 38.46% | 34.78% | - |
| Blood accumulation in pericardial sac | - | `4.35% | 63.16% |
| Malformed ISV/DLAV (includes thin ISV/DLAV) | 83.3% | 29.41% | - |
| Malformed cranial vessels | - | 34.38% | 64.29% |
| Head hemorrhage | 16.38% | 10.87% | 10.53% |
| NaAsO2 (400 ug/mL) exposure time | 0-72 hpf | 24-48 hpf | 48-72 hpf |
| Pericardial edema | 26.32% | 8.77% | - |
| Blood accumulation in pericardial sac | 52.63% | 8.62% | 10% |
| Malformed ISV/DLAV (includes thin ISV/DLAV) | 25% | 3.39% | - |
| Malformed cranial vessels | 50% | 26.53% | 61.54% |
| Head hemorrhage | 5.26% | - | - |
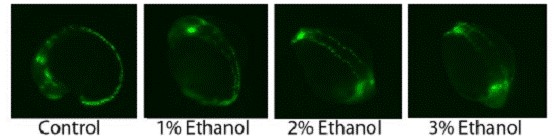
Figure 6. Zebrafish embryos exposed to different concentrations of ethanol from 50% epiboly overnight.
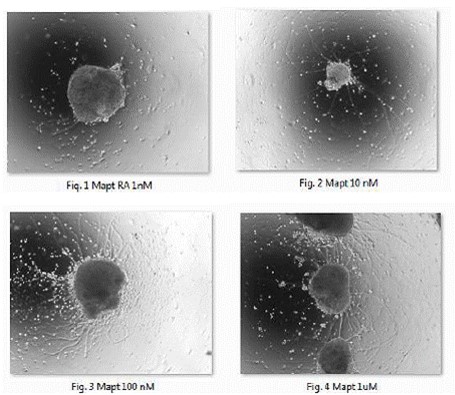
Figure 1-4. Bright-field light micrographs of retinoic acid (RA)-induced neruralization
of embryoid bodies (EBs) in murine microtuble associated protein tau (Mapt) ES cells.
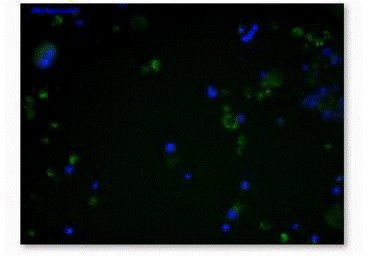
Figure 5. Fluorescence micrograph of Mapt ES cells were
stained with β-Gal and DAPI.
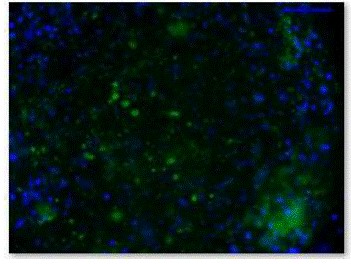
Figure 6. Fluorescence micrograph of β-gal staining of EBs.
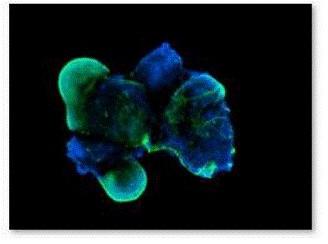
Figure 7. Fluorescence staining of EBs with β-III tubulin
antibody and DAPI
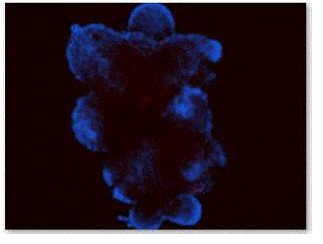
Figure 8. Flourescence staining of EBs with GFAP
antibody and DAPI.
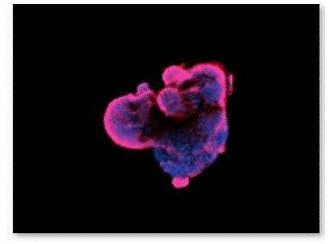
Figure 9. Fluorescence staining of EBs with O4 antibody
and DAPI.
Figure 10. Defferentiation of EBs in to neuronal cells.
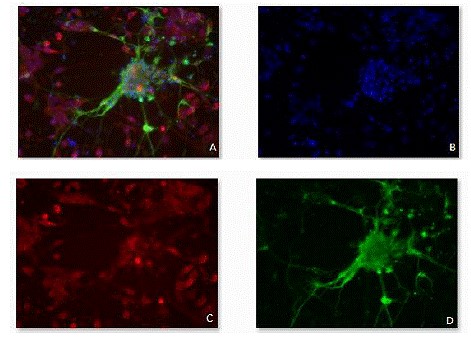
Figure 11. Fluorescence micrographs of differentiated cells stained wiht
antibody markers.
tubulin (left), Q4 antibody (right)] against Syto16 (blue channel) was also demonstrated using
low throughput flow-based system. (Agilent, Bioanalyzer 2000)
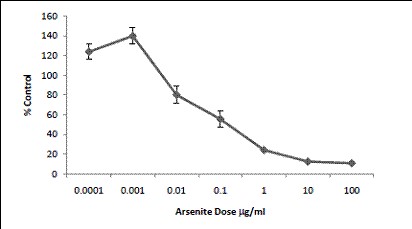
Figure 13. Arsenite (sodium salt) increased the proliferation of cells from
0.1-1hg/mL. At exposures in excess 10mg/mL, a decrease in cell proliferation
was observed.
| Original | Current Orig | EPA | IA1 & IA3 | |
| ABCG2 | Meox1 | Abcg2 | Bmpr2 | Ngn1 |
| Bmp4 | Mycl2 | Mbp4 | Egr1 | tbd |
| Cdh1 | Nodal | Mapt | Gsta2 | tbd |
| Fgf4 | Pcam1 | Nodal | Hmox1 | tbd |
| GATA3 | Pou5f1 | Pou5f1 | Nrf2 | tbd |
| Gsc | Syn1 | tbd | ||
| Mapt | Tie1 | |||
| Mef2a | Wnt3 |
Table 3 shows a list of the origninal proposed 16 gene/clones to be used in the screening assays. Five of the original genes were kept (Orig). Five new genes were selected from data generated by the EPA. Ng1 was selected based on data from IA1, and five genes are to be determined (tbd) to allow for more collaborative research with IA1 and IA3.
Journal Articles: 3 Displayed | Download in RIS Format
| Other center views: | All 13 publications | 3 publications in selected types | All 3 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Bondesson M, Gustafsson J-A. Does consuming isoflavones reduce or increase breast cancer risk? Genome Medicine 2010;2(12):90. |
R834289 (2010) |
Exit Exit Exit |
|
|
Hester SD, Belmonte JM, Gens JS, Clendenon SG, Glazier JA. A multi-cell, multi-scale model of vertebrate segmentation and somite formation. PLoS Computational Biology 2011;7(10):e1002155. |
R834289 (2010) |
Exit Exit Exit |
|
|
Pinto C, Hao R, Grimaldi M, Thrikawala S, Boulahtouf A, Ait-Aissa S, Brion F, Gustaffson J, Balaguer P, Bondesson M. Differential activity of BPA, BPAF and BPC on zebrafish estrogen receptors in vitro and in vivo. TOXICOLOGY AND APPLIED PHARMACOLOGY 2019;380:114709. |
R834289 (2010) |
Exit Exit |
Relevant Websites:
College of Natural Science and Mathematics Exit
Progress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
