Grantee Research Project Results
Final Report: Clinically Relevant IgE-Cross-Reactivity of Nut Allergens
EPA Grant Number: R834066Title: Clinically Relevant IgE-Cross-Reactivity of Nut Allergens
Investigators: Schein, Catherine H. , Teuber, Susanne , Maleki, Soheila
Institution: The University of Texas Medical Branch - Galveston , University of California - Davis , USDA
EPA Project Officer: Aja, Hayley
Project Period: December 1, 2008 through November 30, 2011
Project Amount: $409,927
RFA: Exploratory Investigations in Food Allergy (2007) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
Summary/Accomplishments (Outputs/Outcomes):
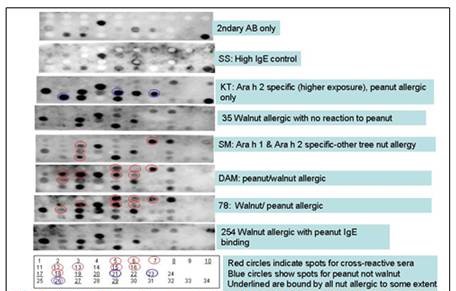
Figure 1. A Spots membrane was developed with controls and several patient sera
to identify peptides that bound IgE from patients with cross-reactivity between
peanuts and walnuts. This membrane retested peptides that had been positive
with at lest one of these sera on a first, larger membrane. The membrane was stripped
and regenerated before each new patient tested (in the order shown here). Sera
from 6 peanut and/or walnut allergic patients were used to identify three different
types of peptides, and specifically a group of peptides that were only recognized
by the cross-reactive patients. Further confirmation of these results, using peptide
microarrays and many more patient sera, would be the goal of a future study.
The complete details of all technical aspects of the project--both negative and positive--the recipient's findings, conclusions, and results, including the associated quality assurance results. The PI and co-investigators were responsible for quality control of distinct project elements:
-Dr. Catherine H. Schein, PI, was responsible for the overall quality assurance (QA) and quality control (QC) aspects of the research and for smooth integration of the computational and experimental aspects of the program. She is an Associate Professor with primary appointment in the Department of Biochemistry and Molecular Biology, with a secondary appointment in Microbiology and Immunology. She is an expert in the sequence and structure of allergenic proteins, was one of the founders of the Structural Database of Allergenic Proteins (SDAP), and is experienced in using the tools constructed for that site. She and her group determined sequences likely to be novel epitopes using the well defined Property Distance (PD) tool that is incorporated in the SDAP, and compared the surface properties of the sequences and structures of epitopes of the nut proteins. She designed the membranes, ordered from a commercial source that had with differing versions of the IgE epitopes to be tested. She also supervised the synthesis of individual peptides for antibody binding experiments and determined their concentration and methods to solubilize them for the studies.
- Serum samples were primarily from the serum bank at UC-Davis, under the direction of Dr. Susanne Teuber, MD, professor of Medicine and expert in tree nut allergies. Dr. Teuber related the clinical phenotype of the patient sera to immunoblotting, and later for correlated this with the results of the individual epitopes detected. The basophil activation assays were also done in Dr. Teuber’s group.
- The immunoblotting and competitive ELISA experiments were carried out in the group of Dr. Soheila Maleki, an expert in peanut allergy and in processing effects on peanut allergenicity. She and her technical assistant used procedures already in place to insure that data were well maintained (by photographing the blots and storing them as a PowerPoint file) and freely available to the other group members.
Conclusions:
- One of the most dangerous and least understood factors in food allergy is how high the degree of sequence or structural similarity must be in order to have a high likelihood of cross-reactivity between food sources. While some have suggested that allergens in foods should have a high percent identity in their amino acid sequences, we have demonstrated that small segments of identity, if these are in an IgE binding area, could lead to cross reactivity.
In this research we chose to compare walnut and peanut allergens, as cross reactivity often is observed between these "nuts", even though they come from quite different botanical sources and their allergenic proteins have a low degree of identity. We used computational methods to identify areas of walnut allergens that were similar in their physicochemical properties and surface exposure to IgE epitopes of peanut proteins. We then made these peptides and showed that certain of the predicted epitopes in walnut allergens bound to IgE in sera of cross-reactive patients as well as previously identified epitopes of peanut. We also found that the cross-reactive sequences tended to be repeats of similar sequences, and that a sequence that was a consensus of these similar repeats detected proteins in many different nut extracts. These last findings suggest that certain determinants of cross reactivity must be ancient sequences that have been maintained in many different tree nuts and seeds perhaps due to their specific properties.
The methods we used can now be applied directly to many different patient sera, using a new experimental method, peptide microarrays. We obtained a great deal of information from these preliminary trials, and found that the SPOTs method gave reproducible results that could be enlarged upon using another method, competitive ELISA. However, we were limited in the number of patient sera we could characterize due to the cost of the SPOTs membranes, and the need to wash and regenerate them between tests of each patient sera. In future studies, assuming we can find additional funding, we plan to use peptide microarrays, where we can test hundreds of different peptides, using sera from hundreds of patients. It is this type of assay that will become the standard for testing the spectrum of allergens a patient reacts to specifically in the next few years.
- The primary benefits of this research are both in limiting exposure to allergens in the environment, and in guidelines for handling patients with severe allergy. Distinguishing allergens from related proteins that present little danger to the environment is a major goal of our research groups. There are many proteins similar to known allergens in all common foods and in other environmental aerosols, such as tree pollens, dust and molds. Simply removing these homologous proteins from all these different sources is not possible, but these results may guide the production of hypoallergenic proteins. However, we can counsel patients to avoid certain foods that they are most likely to react to, based on the presence in their sera of IgEs that recognize common sequences. Similarly, many companies now are using proteins in foods to enhance their properties, or even preparing protein preparations and vaccines for injection into humans. Before these products are approved, it is vital to be able to determine their allergenic potential. We have established methods to compare novel proteins to known allergens, so as to evaluate the risks to the public and to those directly involved in producing or processing them. Here we have further identified specific markers in the sequences of these proteins that might classify them as particularly virulent allergens.
Our promising results, which show that there are certain common areas of allergenic proteins that could mediate cross-reactions, will be most valuable if we are able to verify them in future studies with tests of many more patient sera. We have shown that we can distinguish IgE epitopes that are common to many nuts.
We used protein models for some of the parts of the allergens, and for comparison of peanut and walnut allergen structures in our papers and posters. All protein models for this project either used the ones that are available through the SDAP website ( SDAP 2.0 - Structural Database of Allergenic Proteins) or were done using the MPACK Suite as described in the references.
Model quality was routinely checked with Procheck. The % Identity of the template to the model was given in papers.
References:
Journal Articles on this Report : 6 Displayed | Download in RIS Format
| Other project views: | All 14 publications | 6 publications in selected types | All 6 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Bowen DM, Lewis JA, Lu W, Schein CH. Simplifying complex sequence information: A PCP-consensus protein binds antibodies against all four Dengue serotypes. Vaccine 2012;30(42):6081-6087. |
R834066 (Final) |
Exit |
|
|
Ivanciuc O, Gendel SM, Power TD, Schein CH, Braun W. AllerML: markup language for allergens. Regulatory Toxicology and Pharmacology 2011;60(1):151-160. |
R834066 (Final) R834823 (2011) R834823 (2013) R834823 (Final) |
Exit Exit Exit |
|
|
Maleki SJ, Teuber SS, Cheng H, Chen D, Comstock SS, Ruan S, Schein CH. Computationally predicted IgE epitopes of walnut allergens contribute to cross-reactivity with peanuts. Allergy 2011;66(12):1522-1529. |
R834066 (2010) R834066 (Final) |
Exit |
|
|
Nesbit JB, Hurlburt BK, Schein CH, Cheng H, Wei H, Maleki SJ. Ara h 1 structure is retained after roasting and is important for enhanced binding to IgE. Molecular Nutrition and Food Research 2012;56(11):1739-1747. |
R834066 (Final) R834823 (2012) R834823 (2013) R834823 (Final) |
Exit |
|
|
Schein CH, Ivanciuc O, Midoro-Horiuti T, Goldblum RM, Braun W. An allergen portrait gallery: representative structures and an overview of IgE binding surfaces. Bioinformatics and Biology Insights 2010;4:113-125. |
R834066 (Final) R833137 (Final) |
Exit |
|
|
Schein CH, Bowen DM, Lewis JA, Choi K, Paul A, van der Heden van Noort GJ, Lu W, Filippov DV. Physicochemical property consensus sequences for functional analysis, design of multivalent antigens and targeted antivirals. BMC Bioinformatics 2012;13(Suppl 13):S9. |
R834066 (Final) R834823 (2012) R834823 (2013) R834823 (Final) |
Exit Exit |
Supplemental Keywords:
IgE epitopes, allergen motifs, competitive ELISA, allergen diagnostics, walnut allergenicity, peanut reactivity, B-cell epitope program, SDAPRelevant Websites:
SDAP 2.0 - Structural Database of Allergenic Proteins Exit
Progress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
