Grantee Research Project Results
2010 Progress Report: Clinically Relevant IgE-Cross-Reactivity of Nut Allergens
EPA Grant Number: R834066Title: Clinically Relevant IgE-Cross-Reactivity of Nut Allergens
Investigators: Schein, Catherine H. , Teuber, Susanne , Maleki, Soheila
Institution: The University of Texas Medical Branch - Galveston , University of California - Davis , USDA
EPA Project Officer: Aja, Hayley
Project Period: December 1, 2008 through November 30, 2011
Project Period Covered by this Report: December 1, 2008 through June 30,2010
Project Amount: $409,927
RFA: Exploratory Investigations in Food Allergy (2007) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
Progress Summary:
Table 1. Characteristics of the patient sera.
| | |||||||
| Subject Number | Age of onset (yrs) | Reaction to Peanut |
Reaction to Walnut | ImmunoCAP IgE to Peanut (KU/L) | ImmunoCAP IgE to Walnut (KU/L) | Peanut Proteins detected on WB | Walnut Proteins detected on WB |
|---|---|---|---|---|---|---|---|
| 1. | 1 | Yes, anphx | No | 21.5 | 0.24 | Ara h 1, Ara h 2, Ara h 3, Ara h 6 | Jug r 2, Jug r 4 |
| 2. | 2 | Yes | Yes, anphx | 1.8 | 2.21 | Arha h 1, Ara h 2 Ara h 3 | Jug r 1, Jug r 2, Jug r 4 |
| 3. | 1 | Yes, anphx | Yes, throat swelling, angioedema | >100 | 8.7 | Ara h 1, 2, 3 | Jug r 1, Jug r 2, Jug r 4 |
| 4. | <2 | Yes, anphx | No | 80 | 0.01 | Ara h 2 | none |
| 5. | < 4 | Yes, anphx | No | 23.1 | 0.12 | Ara h 1, Ara h 2, Ara h 6 | none |
| 6. | 2 | No | Yes, anphx | 0.94 | 10.2 | Ara h 1, Ara h 2, Ara h 3, Ara h 6 | Jug r 1 Jug r 2 |
| Spot # Membrane | Allergen name | PD value | Sequence and psition in allergen | ||
|---|---|---|---|---|---|
| 1 | Ara h 2 | 0.00 | 24 | QWELQGDR | 31 known IgE epitope |
| 2 | Jug n 2 | 8.03 | 351 | SFEDQGRR* | 358 |
| 3 | Jug r 2 | 8.21 | 463 | DYEGQGRR* | 470 |
| 4 | Ara h 2 | 0.00 | 27 | DRRCQSQLER | 36 Known IgE epitope |
| 5 | Ara h 2 | 5.23 | 42 | DDQCQRQLQR | 51 |
| 6 | Jug r 2 | 5.87 | 140 | QRQCQQRCER | 149 |
| 7 | Ara h 2 | 7.08 | 96 | EQRCCNELNR | 105 |
| 8 | Jug r 2 | 8.00 | 76 | YEQCQQQCER | 85 |
| 9 | Jug r 2 | 8.12 | 49 | DQRSQEERER | 58 |
| 10 | Jug r 2 | 8.22 | 101 | QRRQQEERER | 110 |
| 11 | Ara h 2 | 8.48 | 94 | DRQMVQHFKR | 103 |
| 12 | Jug r 2 | 8.65 | 532 | QNNIINQLER | 541 |
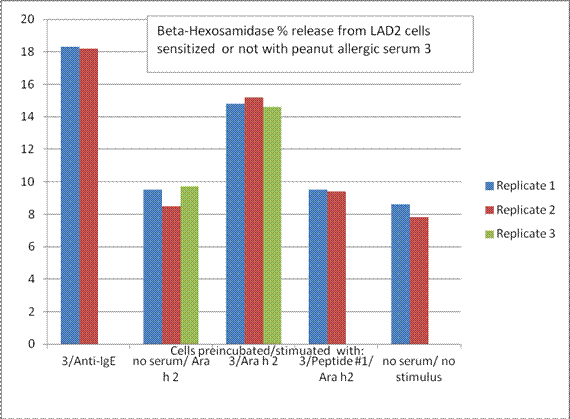
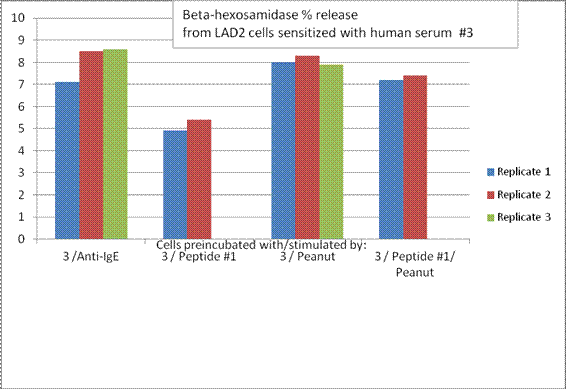
Aim 3 - Summary: The data were observed in terms of the degree of cross reactivity of the patients for peanuts and tree nuts. It was clear that the walnut sensitive individuals recognized sequences selected from walnut allergens better than the patients who were sensitive only to peanuts, or cross-reactive with a mild allergy to walnut. The peptides described in Aim 2 are undergoing evaluation of their ability to inhibit degranulation of a mast cell line and basophil cell line. Originally, the grant had proposed use of human basophils, however, the variability between donors was too high to visualize the slight differences in basophil activation that could be expected by inhibition of only one IgE epitope using the synthesized peptides. There also were difficulties with solubility of the peptides but work was done with the most soluble, peptide 1, as of this progress report. This peptide, a known IgE binding sequence from Ara h 2, could somewhat inhibit degranulation by Ara h 2 from the LAD2 mast cell line. The ability of sera to sensitize basophils or the mast cell line to Ara h 2 also was dependent on recognition of the whole protein in the Western Blots (Figure 1.1).
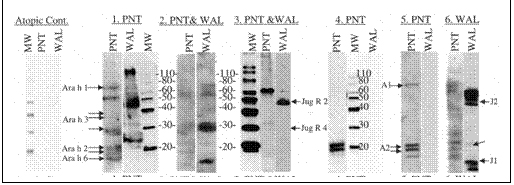
Figure 1.1. Immunoblots of IgE binding by peanut- and/or walnut allergic individuals. Patients
are depicted by nubers (1-6 as above in Table 1) and the clinical allergy to peanut (PNT)
or walnut (WAL). A serum from patient with pollen allergy, MW: protein standard markers.
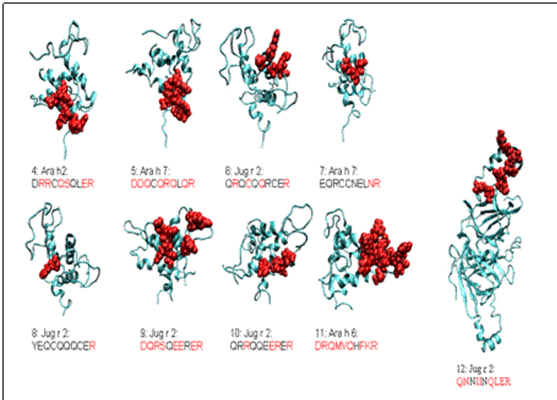
Figure 1.3. The surface exposure of sequences from other nut allergens that were similar (low
PD value) to a known epitope of Ara h 2 (Peptid 4) are plotted on models of allergen structures. The
models, from SDAP or prepared for this study, are shown in ribbon format, with the surface exposed
areas of the potential epitopes shown space filling in red. THe surface exposed residues (>30%
solvent exposure according to GETAREA) in each sequence are highlighted by red type. Novel
epitopes 6,9, and 10 are in the N-terminal extension of Jug r 2, which we modeled for this worki.
Peptid 12, which has a higher PD value (lower similarity) to Peptid 4 than the others and did not
react with IgE in patient sera, is situated in the mature, vicilin region of Jug r 2 (far right figure,
drawn to scale), which has a cupin fold. Peptid PD values are shown in the second part of
Table 2; reactivity in SPOTs in section 1.3.
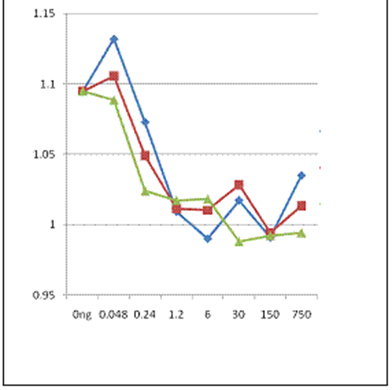
Figure 2.1. Both a peptid of a known epitope of Ara h 2 (DRRCQSQLER,
blue line) and a walnut peptide with low PD to the peanut one
(QRQCQQRCER, orange) can compete with purified, recombinant
ARA h 2 for biding to IgE from patient sera. However, combining the
peptides (green line0 does not add to the inhibition, suggesting both
peptides bind to the same pool of antibodies in the sera.
Future Activities:
(Aim 1) The original characterization was done with 6 patients, and one control selected to have high serum IgE but primary allergy to pollen, rather than nuts. We are expanding the patient pool, and will characterize the new group of sera as below, using strips of selected sequences on spot assays, Western blotting, and, where appropriate, competition assays.
(Aim 2) Other characterized sera will be tested in these ELISA competitions.
(Aim 3) The finding that the Peptide 1, a known IgE binding sequence from Ara h 2, could inhibit basophil degranulation stimulated by Ara h 2, suggests that this might aid in a novel form of de-sensitization (or antibody blocking) therapy using peptides that can bind to the IgE but not trigger degranulation. Dr. Teuber’s group now is trying the other peptides, in combination with different sera, to see how specific the effect is for the chosen peptide.
References:
Journal Articles on this Report : 1 Displayed | Download in RIS Format
| Other project views: | All 14 publications | 6 publications in selected types | All 6 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Maleki SJ, Teuber SS, Cheng H, Chen D, Comstock SS, Ruan S, Schein CH. Computationally predicted IgE epitopes of walnut allergens contribute to cross-reactivity with peanuts. Allergy 2011;66(12):1522-1529. |
R834066 (2010) R834066 (Final) |
Exit |
Progress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
