Grantee Research Project Results
2012 Progress Report: Multi-Sensor Reporter Cell Technology to Assess Hazard Involving Endocrine Signaling Pathways
EPA Grant Number: R835165Title: Multi-Sensor Reporter Cell Technology to Assess Hazard Involving Endocrine Signaling Pathways
Investigators: LeBlanc, Gerald A.
Institution: North Carolina State University
EPA Project Officer: Aja, Hayley
Project Period: March 1, 2012 through February 28, 2016 (Extended to February 28, 2017)
Project Period Covered by this Report: March 1, 2012 through February 28,2013
Project Amount: $950,507
RFA: Developing High-Throughput Assays for Predictive Modeling of Reproductive and Developmental Toxicity Modulated Through the Endocrine System or Pertinent Pathways in Humans and Species Relevant to Ecological Risk Assessment (2011) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
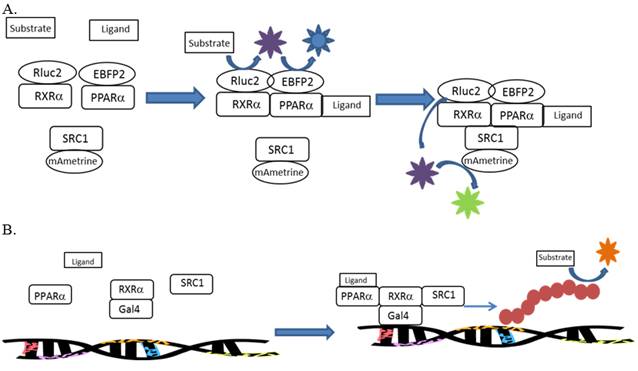
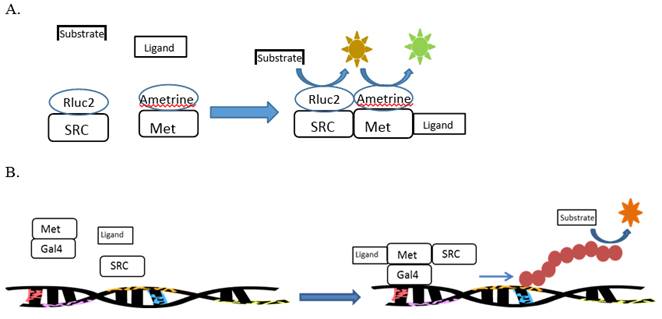
The high-throughput evaluation of toxicity pathways is an emerging paradigm for future toxicity characterization. In order to meet the goals of this paradigm, methods are needed to assess toxicity pathways using as few assays as possible. We propose the use of multi-sensor cell-based reporter assays to meet this need. These assays will utilize both bioluminescence resonance energy transfer (BRET) and reporter gene assays to evaluate interactions of individual chemicals and chemical mixtures along nuclear receptor-mediated signaling pathways. Methods are being developed for the evaluation of chemical effects on the PPARα:RXRα:SRC1 signaling pathway, PPARγ:RXRα:SRC1 signaling pathway, RXRα:RXRα:SRC1 signaling pathway, and the MET:SRC signaling pathway. The former three pathways utilize human receptors that regulate various aspects of glucose and lipid homeostasis. The latter pathway utilizes receptors from the invertebrate Daphnia that regulates various aspects of sex determination and reproductive maturation in arthropods. The development of a high-throughput assay that would identify specific toxicological targets within signaling networks would provide a wealth of mechanistic information on the potential hazard of individual chemicals. This mechanistic data could then be used to model toxicity associated with chemical mixtures.
Progress Summary:
- gal4-RXRα(DEF)-Rluc2
- PPARα(ORF)-EBFP2
- gal4-PPARα(DEF)
- RXRα(ORF)-EBFP2
- SRC1(ORF)-mAmetrine
- PPARγ(ORF)-EBFP2
- gal4-PPARγ(DEF)
- mAmetrine-MET-GAL4
- GAL4-MET-mAmetrine
- Rluc2-SRC
- SRC-SRC-Rluc2
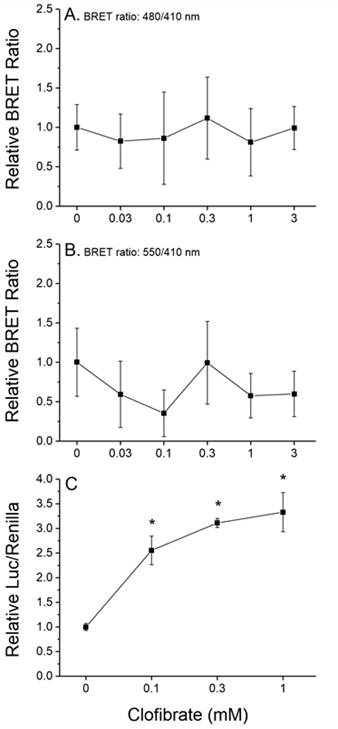
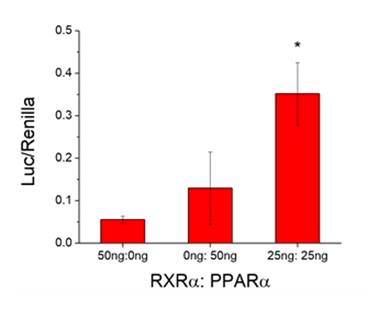
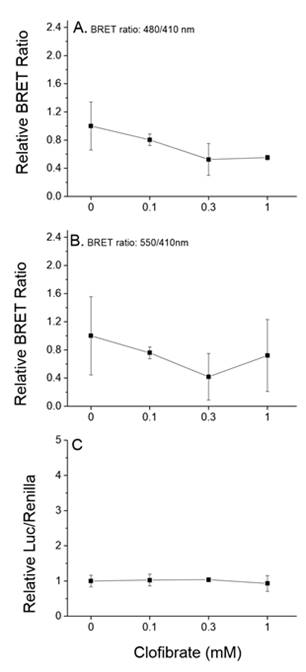
Future Activities:
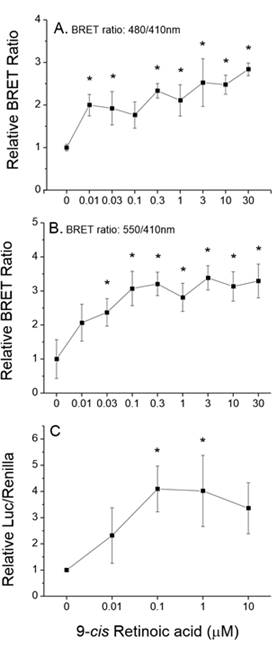
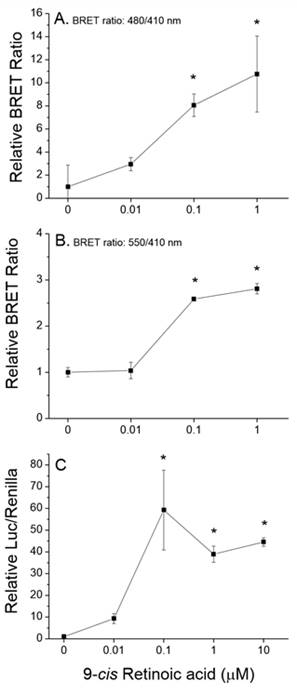
Assays were designed, constructed, and evaluated for the screening of chemicals for activity towards two additional signaling pathways: human PPARα:RXRα:SRC1 and daphnid MET-SRC. These assays will be validated during year 2 of the program.
Journal Articles:
No journal articles submitted with this report: View all 26 publications for this projectSupplemental Keywords:
Multi-sensor reporter cell signaling, signaling pathways, endocrine disruptors, assays, ligandsProgress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
