Grantee Research Project Results
Final Report: A Multi-scale Dose-response Model of AHR Toxicity Pathway Activation in the Human Liver
EPA Grant Number: R835000Title: A Multi-scale Dose-response Model of AHR Toxicity Pathway Activation in the Human Liver
Investigators: Bhattacharya, Sudin
Institution: The Hamner Institutes
EPA Project Officer: Chung, Serena
Project Period: June 1, 2011 through May 30, 2015 (Extended to April 30, 2016)
Project Amount: $750,000
RFA: Computational Toxicology: Biologically-Based Multi-Scale Modeling (2010) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
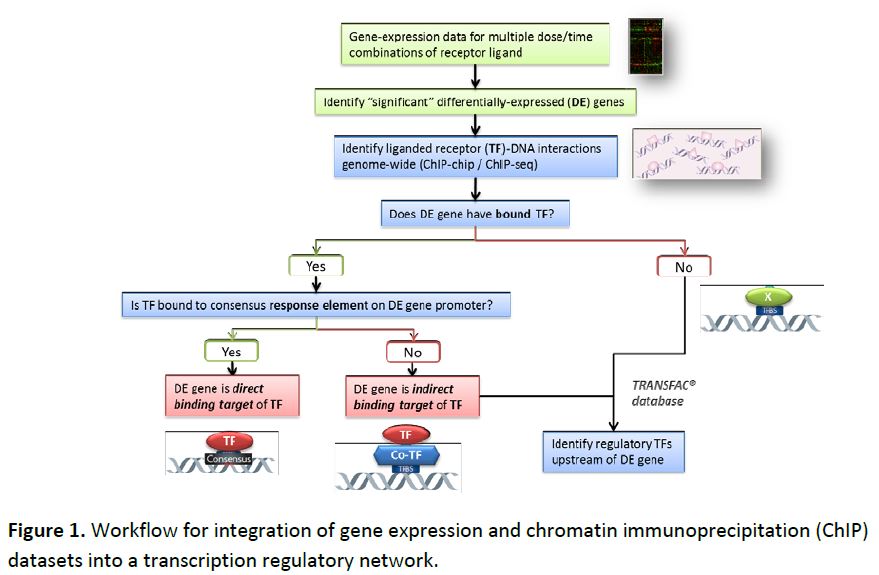
The most significant progress from the work funded in this project has been the development of a computational methodology for integration of varied genomic data sets into transcription regulatory networks underlying toxicity pathways. In the first phase of the project, we applied this approach (Figure 1) to the peroxisome proliferator-activated receptor alpha (PPARα) pathway. Gene expression and genome-wide chromatin immunoprecipitation (ChIP) data sets from primary human hepatocytes were integrated into a functional PPARα network, and analysis and visualization tools were developed for downstream analysis of the phenotypic processes triggered by PPARα activation. This work was published in Chemico-Biological Interactions.
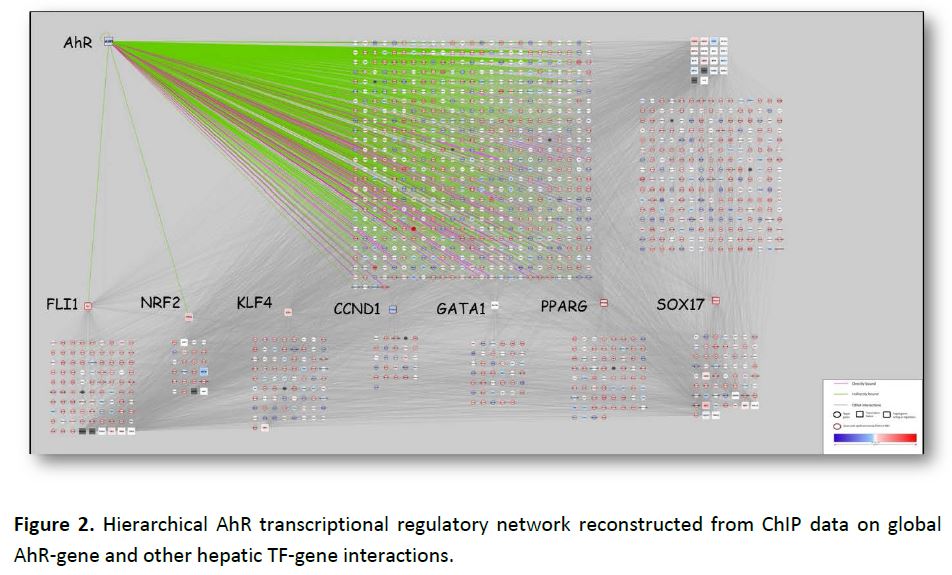
This approach has since been extended to the aryl hydrocarbon receptor (AhR) pathway, where we are analyzing published global AhR ChIP-chip and gene expression data from hepatic tissue of immature ovariectomized mice. These data sets were integrated for reconstruction of the AhR network, along with genome-wide location data for other hepatic transcription factors (TF). AhR ChIP-enriched regions were associated with a target gene if they were located between 10 kb upstream of the transcription start site and the 3’ UTR. Differentially expressed genes were divided into three categories: (i) direct targets, if they were associated with AhR binding at a high-scoring DRE; (ii) indirect targets, if they were associated with AhR binding but not at a high-scoring DRE; and (iii) unbound targets, if they were not associated with AhR binding. The hierarchical transcriptional regulatory network was reconstructed by combining these three groups of genes with AhR-gene and other TF-gene interactions (Figure 2). The network contained a total of 1241 differentially expressed genes and 44 TFs, with AhR, FLI1, NRF2, KLF4, CCND1, GATA1, PPARG and SOX 17 emerging as major transcriptional hubs.
Summary/Accomplishments (Outputs/Outcomes):
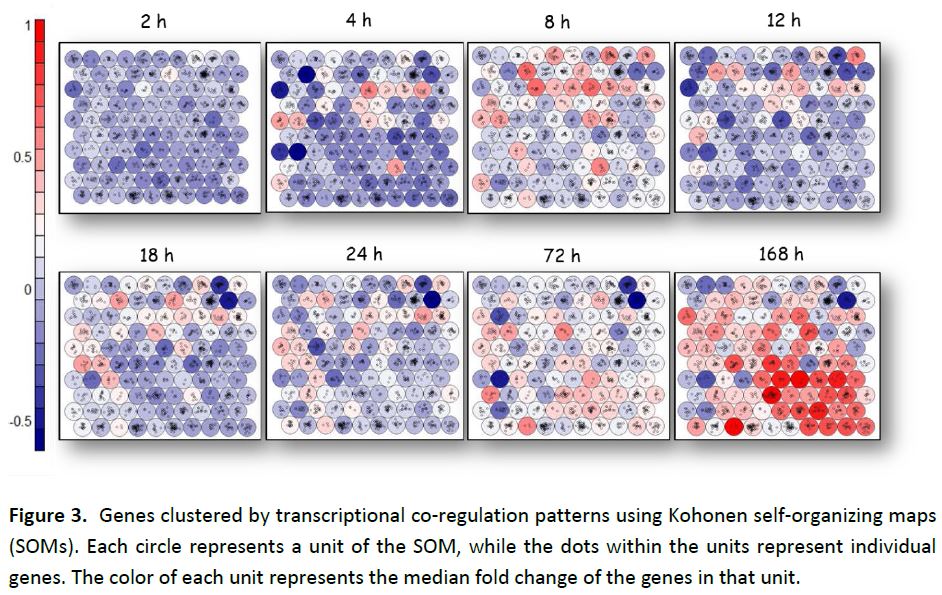
To investigate the pattern of gene expression in groups of co-regulated genes, we classified the genes according to common transcriptional regulators using Kohonen self-organizing maps (SOM). Median gene expression values were then superposed on the units of the SOM (Figure 3). The SOM at 168 h. shows distinct localization of highly expressed genes into adjacent units of the SOM, revealing a relationship between transcriptional co-regulation and gene co-expression.
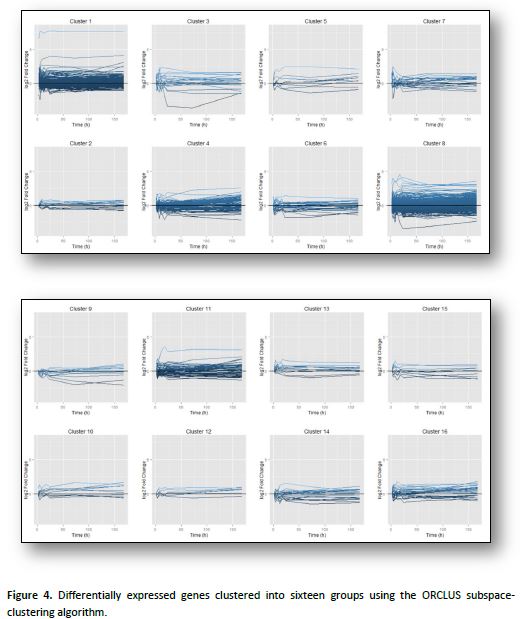
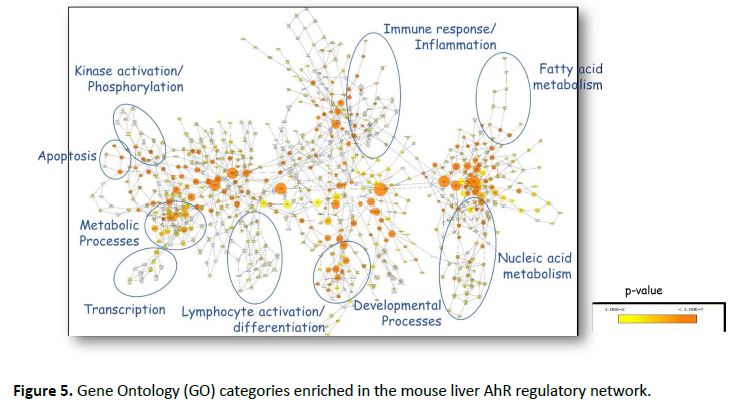
We further investigated the pattern of gene co-expression by using ORCLUS, a subspace-clustering algorithm especially suited to high-dimensional problems with feature localization. This revealed sixteen distinct clusters of genes, grouped by TF-regulation and expression pattern (Figure 4). The LASSO regression algorithm was then used to identify the key regulatory TFs that shape the pattern of expression of each cluster (Table 1). Visualizing the Gene Ontology (GO) categories enriched in the network revealed activation of metabolic, immune and developmental processes (Figure 5). This work is currently being prepared for publication.
In future work, we will use data from recently completed gene expression and AhR ChIP-seq studies of TCDD-treated rat and human primary hepatocytes to assemble AhR hepatic regulatory networks for the human and rat, and carry out a comparative study of the networks in the three species. We will also map the differences in network activation between periportal and centrilobular hepatocytes in the rat liver.
In parallel, we have developed a “virtual tissue” model of the liver lobule in this project. This model was implemented with a modified cluster aggregation algorithm where particles representing endothelial cells underwent random diffusion from the edge of the lobule towards the central vein. The diffusing network of endothelial cells created the sinusoidal vasculature permeating the lobule, which was then used as a framework to add alternating layers of hepatocytes to the lobule (Figure 6A). This virtual liver model was then incorporated into the multi-scale, multicellular modeling environment CompuCell3D (http://www.compucell3d.org/). An intracellular signaling model of Wnt/Beta-catenin and TCDD-induced cytochrome P-450 (CYP) induction was developed in JARNAC (http://sbw.kgi.edu/software/jarnac.htm) and embedded in each hepatocyte in the CompuCell3D lobule model (Figure 6B). This multi-scale model was simulated for various doses of TCDD, exhibiting progression of CYP induction from centrilobular areas at low doses (Figure 7A) to pan-lobular induction at high doses (Figure 7B).
In subsequent steps, we will extend the model to simulate cell proliferation at higher doses of TCDD and link the CompuCell3D model to a PBPK model to estimate hepatic TCDD dose and incorporate AhR network information derived in the previous phase of the work.
We have presented various aspects of this work at multiple meetings including the Society of Toxicology Annual Meeting. We have also published a review article on modeling drug- and chemical- induced hepatotoxicity with systems biology approaches in Frontiers in Physiology, as well as a review book chapter.
Journal Articles on this Report : 2 Displayed | Download in RIS Format
| Other project views: | All 11 publications | 3 publications in selected types | All 2 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Bhattacharya S, Shoda LK, Zhang Q, Woods CG, Howell BA, Siler SQ, Woodhead JL, Yang Y, McMullen P, Watkins PB, Andersen ME. Modeling drug- and chemical-induced hepatotoxicity with systems biology approaches. Frontiers in Physiology 2012;3:462. |
R835000 (2012) R835000 (Final) |
Exit Exit |
|
|
McMullen PD, Bhattacharya S, Woods CG, Sun B, Yarborough K, Ross SM, Miller ME, McBride MT, LeCluyse EL, Clewell RA, Andersen ME. A map of the PPARα transcription regulatory network for primary human hepatocytes. Chemico-Biological Interactions 2014;209:14-24. |
R835000 (2013) R835000 (Final) |
Exit Exit Exit |
Supplemental Keywords:
Genomic data sets, transcription regulatory networks, toxicity pathways, proliferator-activated receptor alpha, PPARα, aryl hydrocarbon receptor, AhR, dose response model, human liver, transcription factorsProgress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
Project Research Results
- 2014 Progress Report
- 2013 Progress Report
- 2012 Progress Report
- 2011 Progress Report
- Original Abstract
2 journal articles for this project