Grantee Research Project Results
Final Report: Differentiating biologically relevant from irrelevant IgE binding to food antigens for improved risk assessment and diagnostic studies using a humanized rat basophil cell line (RBL 30/25)
EPA Grant Number: R834065Title: Differentiating biologically relevant from irrelevant IgE binding to food antigens for improved risk assessment and diagnostic studies using a humanized rat basophil cell line (RBL 30/25)
Investigators: Goodman, Richard E.
Institution: University of Nebraska at Lincoln
EPA Project Officer: Aja, Hayley
Project Period: May 1, 2009 through March 31, 2011 (Extended to April 30, 2012)
Project Amount: $372,340
RFA: Exploratory Investigations in Food Allergy (2007) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
- Develop a standardized basophil activation assay to assess the biological activity of human IgE that is cross-reactive in static binding assays;
- Use the basophil assay to evaluate whether IgE specific to complex carbohydrates on plant glycoproteins is responsible for false positive results;
- Identify biologically relevant cross-active proteins (peptides) of related legume seeds that induce basophil histamine release through allergen specific IgE binding.
Summary/Accomplishments (Outputs/Outcomes):
The primary risk of allergy from a new genetically modified food (GM) crop would be the introduction of a protein that already causes allergies into a food that does not already express the protein (Nordlee, et al., 1996; Goodman, et al., 2008). If the source of the gene is allergenic or the sequence of the protein is even modestly similar to a known allergen, serum IgE tests are required using blood samples from those allergic to the source of the known or similar allergen. However, sometimes IgE binding in the laboratory gives false-positive results (Goodman, et al., 2008; Martens, et al., 2011). Therefore, additional tests are required to prove whether IgE binding to a specific protein is capable of causing symptoms due to activation of mast cells and basophils (Kleine-Tebbe, et al., 2006; Goodman and Tetteh, 2011). Although there are no human cell lines that could mimic natural basophils or mast cells in culture, a rat basophil leukemia cell line (RBL-2H3) has been genetically engineered to express the high affinity human IgE receptor (FcεR1) (Vogel, et al., 2005; Lin, et al., 2007). The humanized RBL 703/21 cells developed by Dr. Lothar Vogel at the Paul Ehrlich Institute (PEI) in Langen, Germany, were donated to us for these studies. Rather than testing histamine release, release of the active enzyme, beta-hexosaminidase, is measured as a less-expensive method of measuring basophil degranulation. The cells are cultured, primed with IgE antibodies from patient sera and finally challenged with suspected allergenic proteins such as peanut allergen, Ara h 2 (Figure 1).
Figure 1. RBL 703/21 cells cultured and sensitized with IgE from patient serum or plasma samples are then challenged with specific allergens or with test proteins or extracts. If significant cross-linking or IgE occurs due to the presence of proteins with two or more IgE binding epitopes, the RBL cells will release β-hexosaminidase enzyme along with histamine and other mediators. The enzyme can be quantitatively measured using substrates that change color upon enzyme catalyzed reaction.
Following transfer of the cells and methods to our lab, our goal was to use the bioassay to test the relevance of IgE binding to cross-reactive carbohydrate determinants (CCD), common to a number of diverse plant proteins (Altmann, 2007) as well as evaluate potential cross-reactivity.
Extracts of commonly consumed legumes (bean family) that represent strong allergens (peanut), moderate allergens (soybean) and relatively non-allergenic food crops (navy beans and peas) were tested. In addition, purified proteins were tested, including major peanut allergen Ara h 2, low- (non-) allergenic peanut agglutinin (PNA); and phytohemmaglutinin (PHA) and alpha-amylase inhibitor (aAI) from common bean. The aAI has been introduced into cowpeas (a different legume) through genetic modification (GM) to control storage beetles. One of the primary questions to test was whether the apparently strong IgE binding to cross-reactive carbohydrate determinants (CCD), specific sugar structures bound to PHA and aAI, can trigger basophil activation and degranulation as an important question for evaluating food safety.
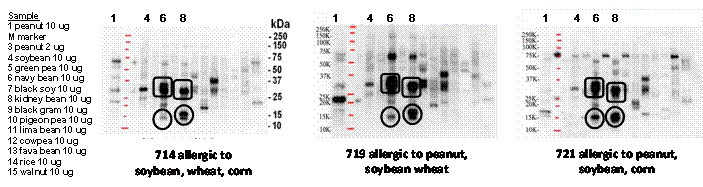
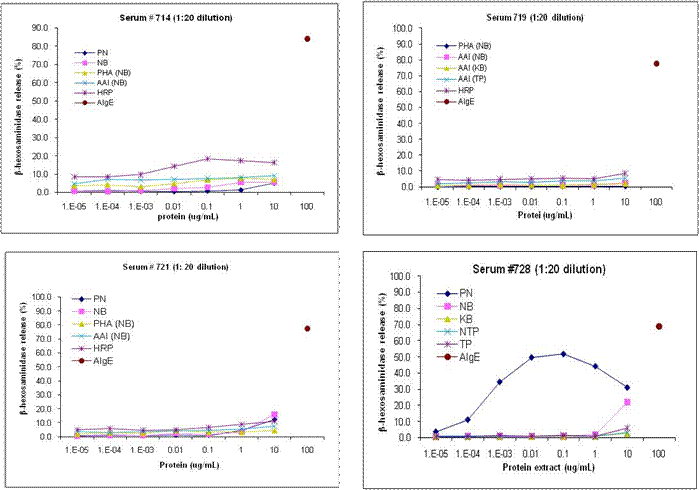
Laboratory IgE binding may be over-predicting risk as serum samples from three subjects with food allergies to peanut and/or soybean indicated, but not to other peas or beans that were tested with extracts of 11 important food legumes shown in Figure 2, below. The IgE binding was primarily to phytohemagglutinin (PHA, in squares) and aAI (circles) of navy bean (lane 5) and kidney bean (lane 7) that are due to IgE binding to CCD. Basophil tests (Figure 3) demonstrated little or no release.
Assay development and evaluation of responses from the subjects in the early part of the studies demonstrated that the assay system is complex and must consider characteristics of the allergic subject, the antigen and cell priming parameters.
Critical parameters:
- RBL 703/21: Cells must be plated at optimum cell density and growth conditions.
- Controls: a) spontaneous release, b) total release, c) pure IgE-anti-IgE release, d) individual allergic serum-anti-IgE, e) non-allergic (or those allergic to other allergens) sera with antigen stimulation help defined appropriate cell status, maximum response, relative concentration of specific IgE vs total IgE for the subject.
- Characteristics of the test protein (purity, structure and identity) must be evaluated.
- Evaluation of in vitro IgE binding under various conditions of antigen (native, denatured, denatured and reduced) is considered along with potential processing of food material.
- Allergic serum/plasma donors should be selected based on clear clinical criteria.
- Serum sample volumes from donors must be sufficient to perform repeated tests.
Figure 3. Release of the mediator β-hexosaminidase from RBL-703/21 cells primed with serum IgE from the four subjects characterized in IgE binding to extracts of peanut, navy bean, kidney bean, GM-aAI (TP) pea and non-transgenic pea figures 3 and 4 [serum 728] demonstrate good release only for peanut extract for the non-CCD binding subject 728. However mediator release for three subjects that show clear IgE binding to PHA and aAI in extracts of navy and kidney beans did not show significant specific release to peanut or navy bean in the RBL assay, and also did not show significant release to PHA or aAI, the major CCD IgE binding glycoproteins. The RBL cells primed with serum from subject 714 did show limited release upon stimulation with horseradish peroxidase (HRP), a highly glycosylated plant protein.
Summary of Findings (Results): We established the RBL 703/21 cells in our laboratory and evaluated stability through multiple rounds of culture splitting as well as freezing. Challenges were performed by harvesting cells from 80-90% confluence, then plating in 96 well culture plates at 100,000 cells per well, followed by the addition of 50 µl of diluted control or allergic sera in complete media, or with standard pure human IgE (at 2 µg/ml in saline) as a positive control. Tests were performed with sera from peanut allergic, peanut and soybean allergic and those allergic to other non-legume sources. Finding appropriately allergic serum donors was a challenge. While peanut allergy affects approximately 1% of the population, a small number, mostly those with coincident asthma, have severe food allergy (Colver, 2006; Sicherer and Sampson, 2007). Although there are multiple allergens in peanut, only two to four “major” allergenic proteins are responsible for most reactions. Selected experiments and data are described below.
- Samples from six peanut allergic subjects and one non-peanut allergic subject with high specific serum IgE to peanut (Table below) were tested using extracts of peanut with the RBL 703/21 cells at concentrations ranging from 0.01 to 10 µg protein per ml peanut extract. Subject (300) who claimed no allergy to foods, but had 100 kU/L peanut specific IgE and 50 kU/L of peanut allergen Ara h 2 (the 2S albumin), released β-hexosaminidase only at high doses of peanut (Figure 4). Subject RG 62, with oral allergy syndrome (mild tingling or itching in the mouth) did not release β-hexosaminidase at any level, yet by immunoblotting bound IgE to major peanut allergens, Ara h 2 and 6. The two highest releasing subjects (297 and 715) reported anaphylaxis to peanut and had very high specific IgE levels to whole peanut extract and to Ara h 2. Subject RGLEG118 reported angioedema to peanut and had substantial release, but at higher doses of peanut (1 and 10 µg/ml) compared to subjects 297 and 715. Subject 728 reactions to peanut were not known, but with high IgE values for peanut (76 kU/L) and Ara h 2 (100 kU/L) did not release β-hexosaminidase at the lowest dose of peanut. These results demonstrated a wide range of concentrations for optimum release that varied remarkably between subjects. SubjectAllergen / symptomsImmunoCAPpeanut (kU/L)ImmunoCAPAra h 2 (kU/L)RG 62Peanut / Oral allergy syndromeNot availableNot Available297Peanut / Anaphylaxis7091299Peanut / Anaphylaxis7489300No food allergy10050715Peanut and soybean / anaphylaxis100100728Peanut / unknown76100RGLEG118Peanut / unknownNot availableNot Available
Note: ImmunoCAP® Phadia (now-Thermo-Scientific) values were measured by clinical diagnostic laboratories using validated in vitro antigen-specific IgE measurements.
Figure 4. RBL 703/21 activation and β-hexosaminidase release stimulated by peanut extract following priming with six peanut-allergic and one non-food allergic subject’s serum or plasma samples. Doses of protein from peanut are shown on the X-axis, percent release based on maximum chemical release is shown on the Y-axis.
- Other tests examined subjects’ serum or plasma for evidence of cross-reactivity between peanut and soybean proteins and peanut and lupine. Clinical data suggest that important cross-reactions of these legumes is relatively rare (Peeters, et al., 2007).
- A study of the safety of GM peas, cowpeas and chickpeas, all transformed with the alpha-amylase inhibitor (aAI) gene, which was cloned from common bean, is under way to consider potential allergenicity using human sera and the humanized RBL assay. These potential GM-products have been under development and evaluation since 1994. In early 2001, Australian regulators suggested to the developer that animal model tests should be performed based on a scientific panel recommendation (FAO/WHO, 2001). The animal model suggested risk (Prescott, et al., 2005). Yet no animal model has been validated to accurately predict allergenicity for humans (Ladics, et al., 2010) and human tests are more likely predictive.
Conclusions:
The humanized rat basophil leukemia cell line 703/21 developed at the PEI in Germany performed well in our hands and provided a reliable biological system to evaluate IgE binding to proteins that are being investigated for potential allergenicity. However, specific parameters must be evaluated for each subject and antigen. There also are no absolute cut-offs for risk assessment, so tests must be based on relative reactivity.
References:
Journal Articles on this Report : 1 Displayed | Download in RIS Format
| Other project views: | All 7 publications | 1 publications in selected types | All 1 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Panda R, Tetteh AO, Pramod SN, Goodman RE. Enzymatic hydrolysis does not reduce the biological reactivity of soybean proteins for all allergic subjects. Journal of Agricultural and Food Chemistry 2015;63(43):9629-9639. |
R834065 (Final) R833135 (Final) |
Exit Exit |
Supplemental Keywords:
mast cell, cross-reactivity, cross-reactive carbohydrate determinants genetically modified, alpha amylase inhibitorProgress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.