Grantee Research Project Results
2019 Progress Report: Center for Integrative Research on Childhood Leukemia and the Environment
EPA Grant Number: R836159Center: Center for Integrative Research on Childhood Leukemia and the Environment - 2015
Center Director: Metayer, Catherine
Title: Center for Integrative Research on Childhood Leukemia and the Environment
Investigators: Metayer, Catherine
Institution: University of California - Berkeley
EPA Project Officer: Callan, Richard
Project Period: September 1, 2015 through August 31, 2019 (Extended to August 31, 2020)
Project Period Covered by this Report: September 1, 2018 through August 31,2019
Project Amount: $2,599,999
RFA: Children's Environmental Health and Disease Prevention Research Centers (2014) RFA Text | Recipients Lists
Research Category: Children's Health , Human Health
Progress Summary:
Overall: During the past year, significant progress has been made to complete the study aims for each Project and Core. In brief, all biospecimens have been obtained, nearly all laboratory assays in Projects 1 and 3 and Core C (Animal model) have been completed, and the majority of laboratory assays for Project 2 have been completed. Core B (Research Translation and Outreach) has completed all of the original aims (with the exception of working with faith groups that ended up not being feasible, yet it was a very small part of the proposed activities). See detailed progress report for each Project and Core, and list of CIRCLE-related publications summarized below.
Core A: Core A continues to support the infrastructure and scientific direction of the Center. We completed the selection and acquisition of the biospecimens that were available at the CA biobank for mother-child cohort specimens, and we have upgraded the data linkage. Another source of biospecimens is the CCLS. Specimens are being used for all three Projects, ensuring sufficient overlap for future integration of the data generated on cytokines (Project 1), exposome and protein adducts (Project 2), and genetic/epigenetic (Project 3).
Aims 1 and 2: Core A continues to support all administrative- and research-related activities in CIRCLE, including working closely with campus post-award unit to monitor the complex book-keeping of 12 chartstrings and 7 sub-awardees split between NIEHS and EPA. The IRB renewal protocol for CIRCLE (#15-09-2232) was approved by the California State Committee for Protection of Human Subjects (CPHS) in April 2019. The UC Berkeley CPHS has received approval for an inter-institutional reliance agreement to the CA State CPHS. The CIRCLE website is updated regularly (http://circle.berkeley.edu/). The Internal Advisory Committee continues to meet every two months to provide updates on all Cores and Projects, and to facilitate scientific interaction between CIRCLE investigators. Minutes are taken to document these discussions.
Core A completed the selection and acquisition of the biospecimens needed for all 3 Projects, and ensure sufficient overlap for future integration of the data generated on cytokines (Project 1), exposome and protein adducts (Project 2), and genetic/epigenetic (Project 3). The Center Director and co-Director were in regular contact with the California Biobank Program Coordinator (Robin Cooley) at the Genetic Disease Program to obtain the archived newborn blood specimens (ANBS) and maternal pregnancy blood samples. We have received all the ANBS from the CCLS participants. Regarding the CA mother-child cohort, we obtained maternal pregnancy blood and platelet samples for 137 cases and ANBS for their corresponding children, as well as maternal and child specimens for 411 controls. The initial linkage between the maternal and child biobanks yielded less samples than expected for cases (which was 200 mother/child pairs for cases). We have received approval from the California Cancer Registry, Vital Statistics, and the CA Biobank to update the linkage of childhood cancers, in an attempt to obtain more sampled for mother/child pairs if needed. This linkage was completed in August of 2019.
Aim 3. Core A continues to support activities in the Community Outreach and Translation Core B, specifically the planning of meetings and calls with the Community Advisory Group (See Details in Core B report). The Center Director, the Pediatric Health Specialist, and other CIRCLE investigators continue to work with Core B leader Mark Miller on various outreach activities to the clinical community. The CIRCLE Director who is also the Chair of the Childhood Leukemia International Consortium (CLIC) organized the 2018 CLIC meeting in Minnesota. This was an opportunity for Dr. Miller to continue to work on research translation with an international group of childhood leukemia researchers. We also facilitate contact with the National Cancer institute (Martha Linet) to help them update the messaging on childhood leukemia prevention.
Aim 4: We held our second External Advisory Committee (EAC) meeting on February 14-15, 2019. The External Advisory Committee provided helpful comments to our research team and sent us a summary report to detail their feedback.
Aim 5: Core A continues to support the professional advancement of Dr. Todd Whitehead, the Career Development Investigator, providing exposure to all scientific and administrative components of research. Dr. Whitehead is working closely with the Leaders of all Cores and Projects. This year, he has continued to facilitate project integration by coordinating the sharing of biospecimens and data between investigators. He is currently working on the analysis of cytokine data (from humans and mice) as part of Project 1. He is also pursuing his own innovative research. Dr. Whitehead was successfully awarded an R24 infrastructure grant in 2017 (with multiple PI Catherine Metayer), where he is leading the work on examining the association between maternal stress during pregnancy and neonatal telomere length. This year, Dr. Whitehead received a TRDRP pilot award to conduct an intervention study designed to reduce exposure to thirdhand smoke in a Chinese-American community in San Francisco. Dr. Whitehead is also cultivating his interest in health communication by developing several innovative research translation materials (at http://circle.berkeley.edu/start-now/ and http://circle.berkeley.edu/dirty-little-secrets/). Additionally, Dr. Whitehead is teaching a graduate level course (Quantitative Exposure Assessment) and has mentored several graduate students in our research group.
Project 1:
Aim 1: We have completed the laboratory work to measure the cytokines in the neonatal biospecimens from the CCLS subjects (~1,000 childhood ALL cases and ~1,000 controls). The following cytokines were measured: IL-1β, IL-4, IL-6, IL-8, IL-10, IL-12p70, GM-CSF, TNF-α, and VEGF. We have completed normalization of the data using the SCONE package in R, and we are currently analyzing the data. Preliminary analyses show that a subset of cytokines may be associated with an increased risk of childhood leukemia. Because the CCLS has well-characterized data on prenatal exposures, we are also conducting comparisons between cytokines and immunotoxic chemicals. Several of the subjects included in the cytokine analyses also overlap with subjects included in Projects 2 and 3, allowing us to integrate these data sources.
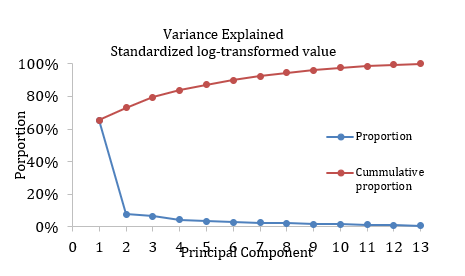
Aim 2: To date, we have obtained maternal pregnancy sera for 128 case mother-child pairs and 476 control mother-child pairs (total n = 1,208) from the California Biobank Program, processed the samples, and analyzed the samples for the following 13 types of immunomodulatory cytokines: IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, GM-CSF, INF-γ, TNF-α, VEGF, and TGF-β1. For samples that yielded a measurement below the detection threshold for a given cytokine, we assigned them a value equivalent to half of the minimum non-zero detected value. For samples measured above the assay saturation, we assigned them a value equivalent to 1.5 times of the maximum detected value. None of the 13 maternal cytokine levels were normally distributed and were standardized and log-transformed for analysis. The maternal cytokine levels are also highly correlated with each other, necessitating dimension reduction techniques such as the principal component analysis. The figure below depicts variance explained by different principal components. We chose a total of four principal components (PCs), which accounted for more than 80% of the overall variance observed in the study sample. The most important contributors to PC1 are IL-1β and TNF-α, while the most important contributors to PC2 are IL-4 and INF-γ (see table below).
Figure 1. variance explained by different principal components
Table 1.The most important contributors to PC1 are IL-1β and TNF-α, while the most important contributors to PC2 are IL-4 and INF-γ
| Standardized log-transformed value | ||||
|---|---|---|---|---|
| Component 1 | Component 2 | Component 3 | Component 4 | |
| IL-1ß | 0.88677 | 0.30885 | -0.0293 | 0.22856 |
| IL-4 | 0.26005 | 0.84225 | -0.257 | 0.27957 |
| IL-5 | ||||
| IL-6 | ||||
| IL-8 | ||||
| IL-12p70 | 0.37387 | 0.42832 | -0.0138 | 0.82045 |
| IFN-γ | 0.37543 | 0.81779 | -0.0249 | 0.19211 |
| TFN-α | 0.88887 | 0.3148 | 0.01376 | 0.20629 |
| VEGF | ||||
| TGF-ß1 | -0.0072 | -0.0269 | 0.99951 | -0.0096 |
| GM-CSF | ||||
We evaluated whether the level of maternal cytokines was associated with child’s year of birth, race/ethnicity, sex, age at the collection of blood spots (in hours), birth weight, gestational age, plurality (singleton vs multiple birth), birth order, mode of delivery (vaginal vs cesarean), mother’s age at delivery, and mother’s birth place (US vs other). These evaluations informed our selection of covariates for the multivariable logistic regression model analyzing the relation of maternal cytokine levels to risk of child acute lymphoblastic leukemia (ALL). We did not observe any significant association between maternal cytokines and ALL risk, when we analyzed each PC as a continuous or dichotomous variable (see Table below). We also categorized the PCs into tertiles and quartiles, and tried to analyze the individual cytokines by themselves (as opposed to part of PCs). None of these analyses yielded any significant findings. Currently, we plan to integrate chemical measurements derived from Project 2 into the analysis and assess whether there is any interaction between maternal cytokines and levels of chemicals present in neonatal blood spots. We note that maternal mid-pregnancy cytokines were only weakly correlated with newborn cytokine levels, which we reported on in last year’s progress report. Newborn cytokine levels as noted on last year’s report, are associated with case-control status in contrast to the maternal cytokines noted below.
Table 2. Newborn cytokine levels as noted on last year’s report, are associated with case-control status in contrast to the maternal cytokines
| Maternal Cytokines and Risk of Childhood ALL | ||||
|---|---|---|---|---|
| Case (%) | Control (%) | OR (95%CI)* | P | |
| Component 1 | 0.96 (0.78-1.18) | 0.68 | ||
| Component 2 | 1.02 (0.83-1.26) | 0.86 | ||
| Component 3 | 0.96 (0.79-1.17) | 0.69 | ||
| Component 4 | 0.97 (0.79-1.20) | 0.81 | ||
| Component 1 | ||||
| Low | 71 (5.55) | 238 (50.0) | 1.00 | |
| High | 57 (44.5) | 238 (250.0) | 0.82 (0.54-1.23) | 0.33 |
| Component 2 | ||||
| Low | 57 (44.5) | 238 (50.0) | 1.00 | |
| High | 71 (55.5) | 238 (50.0) | 1.15 (0.49-1.12) | 0.52 |
| Component 3 | ||||
| Low | 73 (57.0) | 238 (50.0) | 1.00 | |
| High | 55 (43.0) | 238 (50.0) | 0.74 (0.46-1.12) | 0.15 |
| Component 4 | ||||
| Low | 61 (47.7) | 238 (50.0) | 1.00 | |
| High | 67 (52.3) | 238 (50.0) | 1.07 (0.72-1.61) | 0.73 |
*Odds ratios and 95% confidence intervals were derived from multivariable logistic regression models adjusting for age at the collection of blood spots (in hours), birth weight, gestational age, plurality, birth order, mode of delivery, mother’s age at delivery, and mother’s birth place.
Aim 3: We are using a mouse model to elucidate mechanisms by which in utero chemical exposures interact with immune development to initiate childhood ALL. To accomplish Aim 3, we have selected appropriate chemical exposures to be used in the mouse model experiments. To this end, we performed a literature search that characterized the known chemical risk factors for childhood ALL. In collaboration with Core C Leader, Scott Kogan, we ranked a series of putative ALL risk factors using the following criteria: the specificity of epidemiological evidence, the consistency of epidemiological findings, the magnitude of risk estimates in epidemiological studies, the prevalence of exposure, the feasibility of a public health intervention, and the urgency of the public health response. Based on these criteria we have selected four initial chemical exposures of interest to test in the mouse model of ALL: polychlorinated biphenyls (PCBs), exemplified by the Arolclor 1260 mixture; polycyclic aromatic hydrocarbons, exemplified by dibenz[a,h]anthracene; insecticides, exemplified by cypermethrin and permethrin; and tobacco, exemplified by a terry cloth contaminated by aged cigarette smoke (i.e., thirdhand smoke). Progress on the exposures of our leukemia-prone mouse model are detailed in the Progress Report for Core C.
The Core C has returned plasma samples for studies of cytokine changes in mothers and 5 week-old offspring following exposure to dibenz[a,h]anthracene, cypermethrin/permethrin, and thirdhand smoke. There was a total of 32 cytokines measured. We have analyzed the cytokine data by looking at summary statistics, running linear models using one cytokine at a time, creating PCAs to group cytokines (one linear model for each PC), and running MANOVA for multiple outcomes in a single model. In the 2-day pups, we have found the following from our preliminary results: (1) pesticide treatment led to a general suppression of cytokine levels relative to controls, (2) DBA treatment did not cause a significant effect, (3) THS treatment led to a general suppression of cytokine levels relative to controls, and (4) the level of correlation between cytokines was impacted by treatment group. In the 5-week donors, we have found the following: (1) pesticide treatment did not cause a significant effect, (2) DBA treatment led to higher levels of 6 cytokines relative to controls (LIF, Basic-FGF, MIG, MIP2, IL-15, PDGF-BB), (3) THS treatment did not cause a significant effect, and (4) the correlation structure was the same for controls versus all mice overall. In addition, we have been analyzing the correlation between cytokines and immunological markers in mice at 5 weeks. These analyses are ongoing and should be completed in early 2020.
Project 2:
Method development (Aim 1): We adapted metabolomics and adductomics methods for 4.7-mm punches from archived newborn blood specimens (ANBS) and validated them with pilot projects (1-5). This aim has been completed.
Measurement and annotation of omic features (Aim 2): Using the methods from Aim 1, we performed metabolomics and adductomics with archived NBS from 387 childhood cases (339 with ALL and 45 with AML) and matched controls from the CCLS. Matching was based on sex, date of birth and the child’s ethnicity. A total of 869 metabolomic features, mostly lipids and fatty acids, were examined for their associations with ALL and annotated based on matches of accurate masses and MS/MS spectra to database searches (6). Regarding adductomics, a total of 28 HSA adducts were quantitated and compared between ALL and AML cases and controls, including Cys34 sulfoxidation products, Cys34 disulfides with low-molecular-weight thiols, and products of reactive carbonyl species (RCS). In year 04, metabolomics and adductomic analyses were extended to 200 leukemia cases and matched controls from the mother-child cohort. These data are currently being uploaded to the secure server for statistical analyses.
Findings to date (Aims 3 and 4): We compared abundances of 869 small-molecule features in extracts of ANBS punches from 332 children that developed ALL and 324 matched controls from the CCLS (6). Subjects were stratified by early (1-5 y) and late (6-14 y) diagnosis and mutually-exclusive sets of lipids and fatty acids were associated with ALL, including 9 and 19 metabolites in the early- and late-diagnosis groups, respectively. In the late-diagnosis group, several clusters of lipids were either positively or negatively associated with ALL. A prominent cluster contained molecules with 18:2 fatty-acid chains, suggesting that newborn exposure to the essential nutrient, linoleic acid, increased ALL risk. Interestingly, abundances of these 18:2 lipids were greater in infants who were fed formula rather than breast milk (colostrum) and increased with the mother’s pre-pregnancy body mass index.
Although comparisons of adduct levels between leukemia cases and controls found no differences in abundances overall, particular subtypes (i.e., B-cell ALL with t(12;21) and T-cell ALL) had higher abundances of adducts of RCS, suggestive of oxidative stress and lipid peroxidation as potentially causal factors. In addition, a Cys34 adduct of homocysteine, showed consistent discrimination between AML cases and controls with a fold change (cases/controls) of 0.66. Since homocysteine is an important intermediate in the folate-methionine metabolism, this may point to alterations in one-carbon metabolism and epigenetic changes as predictors of AML. We also performed semi-targeted analyses of small molecules chosen a priori as possible biomarkers of childhood ALL, and we detected several targeted molecules that had been selected as putative causes of CL (metabolites of benzene, biomarkers of coffee and alcohol consumption, smoking). However, none of these targeted molecules were detected at significantly higher levels in leukemia cases.
Aim 5: We have completed the metabolomics measurements on 244 maternal/child paired samples (244 neonatal blood spots and 244 maternal serum samples) and are currently conducting adductomic measurements on these samples. The analyses comparing omic features between pairs of ANBS and maternal blood samples collected during pregnancy will be completed in the upcoming months.
Project 3:
Aims 1 and 2: In Year 4 we have continued to analyze DNA methylation data on neonatal blood spots, pre-B cells, and leukemia blasts, and constructed new datasets using DNA methylation arrays (Illumina HM850K). This work has yielded some new and interesting results, and we are wrapping up analyses of environmental variables. We have also participated in PACE Consortium (Pregnancy and Childhood Epigenetics) activities led by NIEHS investigator Stephanie London which involve the meta-analysis of our HM450K methylation data with many other groups also studying neonatal DNA methylation patterns caused by environmental and demographic variations among populations.
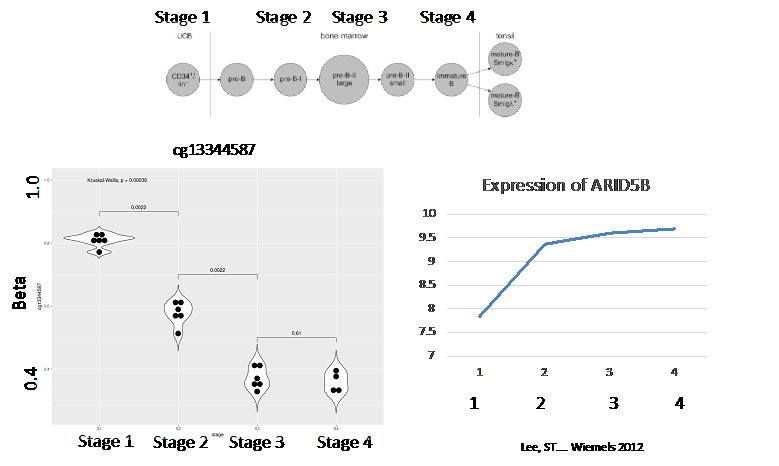
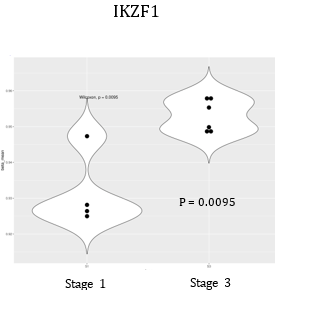
In last year’s progress report, we noted several differentially methylated regions (DMRs) associated with ALL status. We have continued these analyses with our newer HM850K data and found additional replicable loci in individual CpGs in the gene ARID5B; additionally, a series of DMRs (using comb-P and DMRCate algorithms) were discovered including additional loci that pertain to B-cell differentiation pathways. Of the DNA methylation biomarkers at birth that predict future leukemia risk, marks include CpGs within the genes MGMT, HLA-B, ABR, and VTRNA2-1 as we reported in last year’s report. This year, we found a CpG site within a B-cell differentiation gene ARID5B to be associated with leukemia status. This gene was discovered using new HM850K data, and the DNA methylation status of this allele reflected a more mature B-cell status as noted in Figure 1. Our analysis on DNA methylation regions (DMRs) also noted a significant effect from IKZF1, another B-cell differentiation gene. In this case, a higher DNA methylation status was associated with ALL risk, which corroborated with its B-cell differentiation status (Figure 2).
Also, our work with PACE consortium colleagues resulted in manuscripts on various birth factors and DNA methylation patterns. We will use these highly replicated results in understanding the role of DNA methylation alterations that may be affected by such environments and be related to leukemia risk.
Our research group is also heavily involved in a new field called “immunomethylomics,” which is the analysis of blood cell types by their DNA methylation patterns and how this influences biology and disease incidence and progression. Our collaborators have identified a fetal origin set of CpG sites (as opposed to postnatal) and we are using this panel to help “date” the origin of leukemias within our population. We have also found that a more mature B-cell status is reflected by more risk to ALL. This concept is illustrated by individual genes as seen in Figures 1 and 2, and also in the B-cell profile defined by Houseman deconvolution using Bakulski reference cell sets.
Figure 1: (top) ARID5B DNA methylation status though B-cell differentiation. This DNA methylation site is significantly associated with ALL status at birth for disease thorough childhood. DNA methylation is lower at more mature B-cell status, which is corroborated by status of IKZF1.
Figure 2: (below) IKZF1 DNA methylation status is higher at birth in children who later contract ALL, and the higher DNA methylation status corroborates with a later stage B-cell profile. Additional significant loci are shown in the DMRcate + comb-P table.
Table 3. DMRcate + comb-P: HM850K case control analysis
| Gene+ | location | DRMCate FDR | deltabeta | comb-p |
|---|---|---|---|---|
| PRDM8 | Promoter | 7.81E-06 | 1.70 | 41.11E-08 |
| B4GAINT4 | Promoter | 1.57E-07 | -4.82 | 1.44E-07 |
| SLC26A8 | Promoter | 8.23E-06 | -7.23 | 109E-05 |
| RWDD2B | Promoter | 9.14E-05 | -29.24 | 0.00001454 |
| TRIM4 | Promoter | 1.57E-07 | -16.66 | 0.0000159 |
| TRHR | Promoter | 0.0000134222 | 6.67 | 0.00002634 |
| PRKCZ | 1.95E-05 | 5.15 | 0.0004333 | |
| SLC12A4 | Promoter | 0.004098571 | 40.32 | 0.0004494 |
| PRDM6 | 0.000179148 | -11.62 | 0.0005673 | |
| TRFRSF1A | Promoter | 0.000546152 | -23.96 | 0.0009706 |
| ESM1 | Promoter | 0.000188862 | -3.47 | 0.001045 |
| ADGRG1 | 0.00566972 | -6.24 | 0.001075 | |
| SKAP1 | Promoter | 0.000703398 | -13.95 | 0.004408 |
| IKZF1 | Enhancer | 0.0000278124 | 7.99 | 0.004408 |
| ANTXR2 | 0.000433301 | 10.15 | 0.005955 | |
| SLC18A2 | Promoter | 0.006700.87 | 7.12 | 0.007408 |
Aim 3: For Aim 3, we have created DNA methylation datasets for mice exposed prenatally to thirdhand tobacco smoke, pesticides, and polychlorobiphenyls (PCBs). We have analyzed DNA methylation in relation to these exposures and are currently preparing manuscripts that incorporate DNA methylation and cytokine changes in relation to these exposures, and leukemia risk in the particular mouse models.
Core B: Core B continues to be extremely active in translating the findings from CIRCLE and from other relevant research groups (such as the Childhood Leukemia International Consortium CLIC) to various audiences.
The COTC aims to improve pediatric environmental health literacy amongst the clinical audiences by organizing and giving presentations and symposia including at the Pediatric Academic Societies, the annual Children with Cancer UK conference, UCSF Pediatric Malignancies Program, and the International Society for Environmental Epidemiology. In the past year, we have had presentations at a variety of meetings including at the Cancer Free Economy conference and University of Pittsburgh Pediatric Oncology Rounds (Pittsburgh, PA), the US EPA Children’s Health Advisory Committee (Washington, DC), the International Society for Children’s Health and the Environment (Merida, Mexico), UCSF’s large national pediatrics conference “Advances and Controversies in Pediatrics”, at the CDCs Cancer Cluster Guidelines meeting (Atlanta, GA), and the University of Southern California Preventive Medicine Rounds (Los Angeles, CA). Vickie Leonard will present on environmental health in Early Care and Education at the California Association for the Education of Young Children Annual Conference in Santa Clara, as well as at the California Head Start Health Institute in Sacramento, both in April.
The CIRCLE COTC has partnered with the California EPA, Office of Environmental Health Hazard Assessment and the Western States PEHSU to produce a children’s environmental health. Topics have included Environmental Justice (EJ) and Children, Air Pollution and Lifecourse Neurologic Impacts, Climate Change and Children’s Health, and an Update on Research from California- based Children’s Environmental Health Research Centers. Many were the first ever symposium to focus specifically that topic. Though at first glance some topics may seem divergent, they are all relevant to our CIRCLE COTC aims; for example, EJ community exposures of concern are also linked to childhood leukemia risk and Latino children have the highest risk for the disease as well as the greatest likelihood of living in an EJ community. These meeting are well attended in person and via internet and videos of the presentations are posted online.
Dr. Miller also joined with others to create a work group to develop changes to National Cancer Institute (NCI) materials to address the potential roles of environmental exposures in childhood cancer and include actions that might reduce risks. Members included representatives from CDC and NIEHS. The work group developed a revision of the NCI children’s cancer fact sheet that now includes discussion of environmental risk factors and is posted on the NCI website (found at: https://www.cancer.gov/types/childhood-cancers/child-adolescent-cancers-fact-sheet). Dr. Miller and Maria Valenti of our COTC (along with collaborators) contributed a chapter on Environmental Health Literacy of Health Care Providers to a book on environmental health literacy organized by NIEHS and published in 2018.
The Story Of Health e-book continues to be successful. We have continued working with the Western States PEHSU and other partners and in fall 2017 released the fourth chapter of the Story of Health (SOH) multimedia eBook (on infertility and reproductive health). The fifth chapter has been released in October 2019 on Lifecourse Exposures and Cognitive Decline. So far, SOH has had over 10,000 registrations for physician, nurse, and health educator continuing education (CE) credit representing approximately 16,000 hours of CE credits to be awarded by the CDC. We have developed a low literacy version of the SOH, in an innovative comic book format in both English and Spanish, for use in our various anticipated programs. The COTC has produced also an innovative “shadow puppet theater” short video titled “Love in the Time of Toxicants”. This provides an alternative format for introducing the ideas in the Story of Health materials for a general community audience. It is available in English and Spanish and has been well received in the US as well as Mexico (https://wspehsu.ucsf.edu/projects/improving-environmental-health-literacy-young-adults/).
Other recent materials developed by the COTC include two innovative microsites. One, “Carlos and Maria Plan a Family”, features a series of infographics on childhood exposures and risk for childhood leukemia. The other, “Dirty Little Secrets about House Dust”, examines household exposures of contaminants attached to dust particles and actions to limit children’s exposure. A series of five engaging infographics raising awareness of risk factors for childhood leukemia were produced in the past year. In addition, a newsletter designed to sustain interest generated by our workshops for parents, educators, and Promotores de Salud was established with the first two issues distributed so far.
The materials described above were released and are available in Spanish and specifically address the needs of the Latino community. They have been used in numerous workshops presented to parents, Promotores de Salud (lay health workers), childcare workers, and at health fairs. Our materials our being used in programs along the border in Texas by promotora and medical and nursing student training sponsored by the El Paso based PEHSU.
Dr. Miller has served during 2018-19 on an expert panel for the CDC on Cancer Cluster Guidelines. He has also been serving the US EPA as a member of the Federal Advisory Committee, the Children’s Health Protection Advisory Committee. Vickie Leonard, RN, PhD has participated in the Environmental Health Subgroup of the California Child Care Health and Safety Work Group. Vickie Leonard presented on why environmental health matters in early care and education at the Stakeholders Roundtable on Safe Siting of Child Care Facilities put on by the Site Assessment Section, Environmental Health Investigations Branch, California Department of Public Health.
The COTC has established a distinguished community advisory group has held annual meetings. Representatives include a health educator from the California Department of Public Health, a community activist from an environmental justice community along the Mexican border, a physician member of the Kaiser Hispanic physicians organization, a member of a non-profit that funds research in childhood leukemia as well as parent and community outreach (who is also a parent of a child with leukemia), an NGO staffer who targets education of the public on risk factors and preventive activities related to breast cancer, and a leader of another Children’s center COTC with extensive experience working with the Latino farmworker community. The Community Advisory Committee has provided helpful direction and feedback on our outreach and materials development and volunteered to help in a variety of concrete ways. The advisory group held its final meeting this year.
Core C: As discussed in our prior progress report, we adapted our experimental exposure design in response to observations made in the course of our studies. In the current project period we made use of Cdkn2a null animals (which are susceptible to leukemia/lymphoma and other tumors) along with bone marrow transplantation (which is permissive for leukemia/lymphoma but limits the development of non-hematopoietic tumors). We have performed the following studies and our analyses to date are summarized.
Third Hand Smoke (THS): Third hand smoke has recently been recognized as an important mechanism through which children are exposed to tobacco carcinogens. We hypothesized that exposure to THS during pregnancy and following birth would contribute to ALL in our mouse models. Exposure regimen consisted of exposing pregnant dams from time of mating until weaning of pups to THS-exposed or control cotton terrycloth. Sera of 2 day old pups was compared between THS-exposed and control-exposed cohorts. These data show that pre-natal exposure to THS suppresses the expression of multiple cytokines/immmunomodulators. Sera of 5 weeks old animals were also compared, which showed normalization of most cytokine levels by week 5, with a persistent deficit in IL-2. Also at 5 weeks of age, we observed that bone marrow and splenic B-cells were reduced in exposed mice. Recipients of bone marrow from exposed mice were followed for disease. Unexpectedly, recipients of bone marrow from THS-exposed animals developed disease more slowly than recipients of bone marrow from control-exposed animals. This result highlights that tobacco smoke exposure can have pleomorphic effects, and could interfere with B-cell development in a manner that suppresses rather than facilitates leukemogenesis. Such a finding might only apply to particular genetic backgrounds and particular predisposing lesions. Further, it might explain prior meta-analyses of human data that show a substantial impact of human maternal smoking during pregnancy on DNA methylation in newborns, but no overall substantial effect on leukemogenesis. Analyses of methylation changes brought by prenatal and early life tobacco smoke exposure are ongoing, as part of the above investigations. Further studies of the impact of tobacco smoke in alternative mouse models of ALL may delineate whether such exposure has neutral or leukemia-promoting effects in other systems.
Polycyclic aromatic Hydrocarbons (PAHs): For the PAH Dibenz[a,h]anthracene we used an exposure protocol of 3 mg/kg exposure every other day during pregnancy for a total of 5 doses. Although analyses are ongoing, to date, analyses of cytokine levels, changes to immune cell numbers and leukemia/lymphoma development have been essentially negative (with no statistically significant differences between control-exposed and PAH-exposed animals.
Insecticides: Cypermethrin and permethrin dosing regimen was refined based on our experience to include a combination of 35 mg/kg cypermethrin + 200 mg/kg permethrin each day for a total of 10 days during pregnancy. This exposure to commercially available pesticide preparations was observed to impact cytokine/immunomodulator serum levels in 2 days old pups and, to cause modest persistent immune changes at 5 weeks of age, but not to influence the development of leukemia/lymphoma in the selected model system. Analyses of methylation changes wrought by prenatal pesticide exposures are ongoing.
At present, we are continuing to follow a number of animals in our PAH and insecticide cohorts for the development of disease, and to analyze our data, including in comparison to results in human studies from other areas of our CIRCLE collaborations.
CIRCLE Data Integration: While waiting for all newly generated data from the three Projects (cytokines, methylation data, and metabolomics/adductomic data) for both study populations (the mother-child cohort and CCLS), we have designed a query database system to efficiently assess availability of omic data and existing exposure data (from questionnaire data and household exposure measurements).
As a proof-of-principle for data integration, we are examining the relationships between the small molecules/protein adducts that we have measured in neonatal blood spots (Project 2, Aim 3) and neonatal immune status, among a small group of participants with existing CCLS pilot data on cytokines (Project 1; N=50). Intriguingly, we observe statistically significant levels of correlation between levels of specific small molecules/protein adducts and concentrations of IL-10/IL-12 in archived neonatal blood spots. Likewise we have identified potential relationships between levels of PCBs measured in settled dust and concentrations of IL-10/IL-12 in archived neonatal blood spots. We are also developing follow-up studies to integrate omic data related to the one-carbon (folate) metabolism pathway. We will work with biostatisticians to adapt statistical methods such as machine learning and weighted quantile sum approaches to our multidimensional data. These analyses will be conducted in calendar years 2019 and 2020.
Journal Articles: 35 Displayed | Download in RIS Format
| Other center views: | All 37 publications | 35 publications in selected types | All 35 journal articles |
|---|
| Type | Citation | ||
|---|---|---|---|
|
|
Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, Herbstman J, Holland N, LaSalle JM, Schmidt R, Yousefi P, Perera F, Joubert BR, Wiemels J, Taylor M, Yang IV, Chen R, Hew KM, Freeland DM, Miller R, Murphy SK. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies:the Children's Environmental Health and Disease Prevention Research Center's Epigenetics Working Group. Environmental Health Perspectives 2017;125(4):511-526. |
R836159 (2018) R834515 (Final) R835436 (2017) |
|
|
|
de Smith AJ, Walsh KM, Morimoto LM, Francis SS, Gonseth S, Jeon S, Chen M, Sun H, Luna-Fineman S, Antillon F, Hansen HM, Kang AY, Smirnov I, Xiao X, Whitehead TP, Barcellos LF, Sender L, Healy J, Laverdière C, Sinnett D, Taub JW, Birch JM, Thompson P, Pombo-de-Oliveira MS, Spector LG , DeWan AT, Mueller BA, Chiang C, Metayer C, Ma X, Wiemels JL (In Press) Heritable variation at the chromosome 21 gene ERG is associated with acute lymphoblastic leukemia risk in children with and without Down syndrome. Leukemia. |
R836159 (2019) |
Exit |
|
|
de Smith AJ, Kaur M, Gonseth S, Endicott A, Selvin S, Zhang L, Roy R, Shao X, Hansen HM, Kang AY, Walsh KM, Dahl GV, McKean-Cowdin R, Metayer C, Wiemels JL. Correlates of prenatal and early-life tobacco smoke exposure and frequency of common gene deletions in childhood acute lymphoblastic leukemia. Cancer Research 2017;77(7):1674-1683. |
R836159 (2017) R836159 (2018) R836159C003 (2017) |
Exit Exit Exit |
|
|
Edmands WMB, Petrick LM, Barupal DK, Scalbert A, Wilson MJ, Wickliffe JK, Rappaport SM. compMS2Miner:an automatable metabolite identification, visualization, and data-sharing R package for high-resolution LC-MS data sets. Analytical Chemistry 2017;89(7):3919-3928. |
R836159 (2017) R836159C002 (2017) |
Exit Exit Exit |
|
|
Felix JF, Joubert BR, Baccarelli AA, Sharp GC, Almqvist C, Annesi-Maesano I, Arshad H, Baiz N, Bakermans-Kranenburg MJ, Bakulski KM, Binder EB, Bouchard L, Breton CV, Brunekreef B, Brunst KJ, Burchard EG, Bustamante M, Chatzi L, Cheng Munthe-Kaas M, Corpeleijn E, Czamara D, Dabelea D, Davey Smith G, De Boever P, Duijts L, Dwyer T, Eng C, Eskenazi B, Everson TM, Falahi F, Fallin MD, Farchi S, Fernandez MF, Gao L, Gaunt TR, Ghantous A, Gillman MW, Gonseth S, Grote V, Gruzieva O, Haberg SE. Cohort profile: Pregnancy And Childhood Epigenetics (PACE) Consortium. International Journal of Epidemiology 2018;47(1):22-23u. |
R836159 (2018) R836159 (Final) R835442 (2018) |
Exit Exit Exit |
|
|
Giddings BM, Whitehead TP, Metayer C, Miller MD. Childhood leukemia incidence in California: high and rising in the Hispanic population. Cancer 2016;122(18):2867-2875. |
R836159 (2017) R836159 (2018) |
Exit Exit Exit |
|
|
Gonseth S, Roy R, Houseman EA, de Smith AJ, Zhou M, Lee ST, Nussle S, Singer AW, Wrensch MR, Metayer C, Wiemels JL. Periconceptional folate consumption is associated with neonatal DNA methylation modifications in neural crest regulatory and cancer development genes. Epigenetics 2015;10(12):1166-1176. |
R836159 (2017) R836159 (2018) R836159 (2019) R836159C003 (2016) R834511 (2014) |
Exit Exit Exit |
|
|
Gonseth S, de Smith AJ, Roy R, Zhou M, Lee S-T, Shao X, Ohja J, Wrensch MR, Walsh KM, Metayer C, Wiemels JL. Genetic contribution to variation in DNA methylation at maternal smoking-sensitive loci in exposed neonates. Epigenetics 2016;11(9):664-673. |
R836159 (2017) R836159 (2018) R836159C003 (2017) |
Exit Exit |
|
|
Kaur M, de Smith AJ, Selvin S, Zhang L, Cunningham M, Kang MW, Hansen HM, Cooper RM, McKean-Cowdin R, Wiemels JL, Metayer C. Tobacco smoke and Ras mutations among Latino and non-Latino children with acute lymphoblastic leukemia. Archives of Medical Research 2016;47(8):677-683. |
R836159 (2018) |
Exit Exit |
|
|
McCanlies EC, Fekedulegn D, Mnatsakanova A, Burchfiel CM, Sanderson WT, Charles LE, Hertz-Picciotto I. Parental occupational exposures and autism spectrum disorder. Journal of Autism and Developmental Disorders 2012;42(11):2323-2334. |
R836159 (2019) R833292 (2012) R833292 (Final) |
Exit |
|
|
Metayer C, Dahl G, Wiemels J, Miller M. Childhood leukemia: a preventable disease. Pediatrics 2016;138 (Suppl 1):S45-S55. |
R836159 (2017) R836159 (2018) |
Exit Exit Exit |
|
|
Miller MD, Valenti M, Schettler T, Tencza B. A multimedia e-book — A story of health: filling a gap in environmental health literacy for health professionals. Environmental Health Perspectives 2016;124(8):A133-A136. |
R836159 (2017) R836159 (2018) |
|
|
|
Miller MD, Valenti M, Schettler T, Tencza B. A story of health: filling a gap in environmental health literacy for health professionals. San Francisco Medicine, Journal of San Francisco Medical Society 2016;89(10):20-24 (Reprinted and edited with permission from Environmental Health Perspective). |
R836159 (2018) |
Exit |
|
|
Milne E, Greenop KR, Petridou E, Bailey HD, Orsi L, Kang AY, Baka M, Bonaventure A, Kourti M, Metayer C, Clavel J. Maternal consumption of coffee and tea during pregnancy and risk of childhood ALL: a pooled analysis from the Childhood Leukemia International Consortium. Cancer Causes & Control 2018;29(6):539-550. |
R836159 (2018) R836159 (2019) |
Exit Exit |
|
|
Orsi L, Magnani C, Petridou ET, Dockerty JD, Metayer C, Milne E, Bailey HD, Dessypris N, Kang AY, Wesseling C, Infante-Rivard C, Wunsch-Filho V, Mora AM, Spector LG, Clavel J. Living on a farm, contact with farm animals and pets, and childhood acute lymphoblastic leukemia: pooled and meta-analyses from the Childhood Leukemia International Consortium. Cancer Medicine 2018;7(6):2665-2681. |
R836159 (2018) R836159 (2019) |
Exit Exit Exit |
|
|
Petrick L, Edmands W, Schiffman C, Grigoryan H, Perttula K, Yano Y, Dudoit S, Whitehead T, Metayer C, Rappaport S. An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies. Metabolomics 2017;13(3):27 (19 pp.). |
R836159 (2017) R836159 (2018) R836159C002 (2017) |
Exit Exit |
|
|
Petridou ET, Georgakis MK, Erdmann F, Ma X, Heck JE, Auvinen A, Mueller BA, Spector LG, Roman E, Metayer C, Magnani C, Pombo-de-Oliveira MS, Ezzat S, Scheurer ME, Mora AM, Dockerty JD, Hansen J, Kang AY, Wang R, Doody DR, Kane E, Rashed WM, Dessypris N, Schüz J, Infante-Rivard C, Skalkidou A. Advanced parental age as risk factor for childhood acute lymphoblastic leukemia: results from studies of the Childhood Leukemia International Consortium. European Journal of Epidemiology 2018;33(10):965-976. |
R836159 (2018) R836159 (2019) |
Exit Exit |
|
|
Rappaport, S.M. Redefining environmental exposure for disease etiology. NPJ Systems Biology and Application. 2018(4):30. |
R836159 (2019) |
Exit |
|
|
Sharp GC, Salas LA, Monnereau C, Allard C, Yousefi P, Everson TM, Bohlin J, XuZ, Huang RC, Reese SE, Xu CJ, Baiz N, Hoyo C, Agha G, Roy R, Holloway JW, Ghantous A, Merid SK, Bakulski KM, Kupers LK, Zhang H, Richmond RC, Page CM, Duijts L, Lie RT, Melton PE, Vonk JM, Nohr EA, Williams-DeVane C, Huen K, Rifas-Shiman SL, Ruiz-Arenas C, Gonseth S, Rezwan FI, Herceg Z, Ekstrom S, Croen L, Falahi F, Perron P, Karagas MR, Quraishi BM, Suderman M, Magnus MC, Jaddoe VWV, Taylor JA, Anderson D, Zhao S, Smit HA, Josey MJ, Bradman A, Baccarelli AA, Bustamante M, Haberg SE, Pershagen G, Hertz-Picciotto I, Newschaffer C, Corpeleijn E, Bouchard L, Lawlor DA, Maguire RL, Barcellos LF, Davey Smith G, Eskenazi B, Karmaus W, Marsit CJ, Hivert MF, Snieder H, Fallin MD, Melen E, Munthe-Kaas MC, Arshad H, Wiemels JL, Annesi-Maesano I, Vrijheid M, Oken E, Holland N, Murphy SK, Sorensen TIA, Koppelman GH, Newnham JP, Wilcox AJ, Nystad W, London SJ, Felix JF, Relton CL. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Human Molecular Genetics 2017;26(20):4067-4085. |
R836159 (2018) R835442 (2018) |
Exit Exit Exit |
|
|
Wallace AD, Francis SS, Shao X, de Smith AJ, Walsh KM, Mckean-Cowdin R, Ma X, Dahl G, Barcellos LF, Wiemels JL, Metayer C. A germ-line deletion of APOBEC3B does not contribute to subtype-specific childhood acute lymphoblastic leukemia etiology. Haematologica 2018;103(1):e29-e31. |
R836159 (2018) |
Exit Exit Exit |
|
|
Wallace AD, Francis SS, Ma X, McKean-Cowdin R, Selvin S, Whitehead TP, Barcellos LF, Kang AY, Morimoto L, Moore TB, Wiemels JL, Metayer C. Allergies and childhood acute lymphoblastic leukemia: a case-control study and meta-analysis. Cancer Epidemiology, Biomarkers and Prevention 2018;27(10):1142-1150. |
R836159 (2018) R836159 (2019) |
Exit |
|
|
Walsh KM, Whitehead TP, de Smith AJ, Smirnov IV, Park M, Endicott AA, Francis SS, Codd V, ENGAGE Consortium Telomere Group, Samani NJ, Metayer C, Wiemels JL. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis 2016;37(6):576-582. |
R836159 (2018) |
Exit Exit Exit |
|
|
Wang R, Wiemels JL, Metayer C, Morimoto L, Francis SS, Kadan-Lottick N, DeWan AT, Zhang Y, Ma X. Cesarean section and risk of childhood acute lymphoblastic leukemia in a population-based, record-linkage study in California. American Journal of Epidemiology 2017;185(2):96-105. |
R836159 (2017) R836159 (2018) R836159C001 (2017) |
Exit Exit Exit |
|
|
Wang R, Metayer C, Morimoto L, Wiemels JL, Yang J, DeWan AT, Kang A, Ma X. Parental age and risk of pediatric cancer in the offspring: a population-based record-linkage study in California. American Journal of Epidemiology 2017;186(7):843-856. |
R836159 (2018) |
Exit Exit Exit |
|
|
Whitehead TP, Metayer C, Wiemels JL, Singer AW, Miller MD. Childhood leukemia and primary prevention. Current Problems in Pediatric and Adolescent Health Care 2016;46(10):317-352. |
R836159 (2017) R836159 (2018) |
Exit Exit Exit |
|
|
Wiemels JL, Walsh KM, de Smith AJ, Metayer C, Gonseth S, Hansen HM, Francis SS, Ojha J, Smirnov I, Barcellos L, Xiao X, Morimoto L, McKean-Cowdin R, Wang R, Yu H, Hoh J, DeWan AT, Ma X. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nature Communications 2018;9(1):286 (8 pp.). |
R836159 (2018) |
Exit Exit Exit |
|
|
Yano Y, Grigoryan H, Schiffman C, Edmands W, Petrick L, Hall K, Whitehead TP, Metayer C, Dudoit S, Rappaport SM. Untargeted adductomics of cys34 modifications to human serum albumin in newborn dried Blood Spots. Analytical and Bioanalytical Chemistry 2019. |
R836159 (2019) |
Exit |
|
|
Schiffman C, Petrick L, Perttula K, Yano Y, Carlsson H, Whitehead TP, Metayer C, Hayes J, Edmands W, Rappaport SM, Dudoit S. Data-adaptive pipeline for filtering and normalizing metabolomics data. Biorxiv. 2018. |
R836159 (2019) |
Exit Exit Exit |
|
|
de Smith AJ, Walsh KM, Francis SS, Zhang C, Hansen HM, Smirnov I, Morimoto L, Whitehead TP, Kang A, Shao X, Barcellos LF, McKean-Cowdin R, Zhang L, Fu C, Wang R, Yu H, Hoh J, Dewan AT, Metayer C, Ma X, Wiemels JL. BMI1 enhancer polymorphism underlies chromosome 10p12.31 association with childhood acute lymphoblastic leukemia. Int J Cancer 2018;143(11):2647-2658. |
R836159 (2019) |
Exit |
|
|
Panagopoulou P, Skalkidou A, Marcotte E, Erdmann F, Ma X, Heck JE, Auvinen A, Mueller BA, Spector LG, Roman E, Metayer C, Magnani C, Pombo-de-Oliveira MS, Scheurer ME, Mora AM, Dockerty JD, Hansen J, Kang AY, Wang R, Doody DR, Kane E, Schüz J, Christodoulakis C, Ntzani E, Petridou ET. Parental age and the risk of childhood acute myeloid leukemia:results from the Childhood Leukemia International Consortium. Journal of Cancer Epidemiology. 2019(59):158-165. |
R836159 (2019) |
Exit |
|
|
Schiffman C, Petrick L, Perttula K, Yano Y, Carlsson H, Whitehead T, Metayer C, Hayes J, Rappaport S, Dudoit S. Filtering procedures for untargeted LC-MS metabolomics data. BMC Bioinformatics. 2019(1):20:334. |
R836159 (2019) |
Exit |
|
|
Petrick LM, Schiffman C, Edmands WMB, Yano Y, Perttula K, Whitehead T, Metayer C, Wheelock CE, Arora M, Grigoryan H, Carlsson H, Dudoit S, Rappaport SM. Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatric acute lymphoblastic leukemia and potential effects of early-life nutrition. Cancer Letters 2019;452:71-78. |
R836159 (2019) |
Exit Exit |
|
|
Yano Y, Schiffman C, Grigoryan H, Hayes J, Edmands W, Petrick L, Whitehead T, Metayer C, Dudoit S, Rappaport S. Untargeted adductomics of newborn dried blood spots identifies modifications to juman serum albumin associated with childhood leukemia. Leukemia Research. 2020(8):106268 |
R836159 (2019) |
Exit |
|
|
Karalexi MA, Dessypris N, Clavel J, Metayer C, Erdmann F, Orsi L, Kang AY, Schüz J, Bonaventure A, Greenop KR, Milne E, Petridou ET. Coffee and tea consumption during pregnancy and risk of childhood acute myeloid leukemia:A Childhood Leukemia International Consortium (CLIC) study. Cancer Epidemiology. 2019(62):101581 |
R836159 (2019) |
Exit |
|
|
Nielsen AB, Zhou M, de Smith AJ, Want R, McCoy L, Hansen H, Morimoto L, Gronbaech K, Johansen C, Kogan SC, Metayer C, Bracci PM, Ma X, Wiemels JL. Increased neonatal level of arginase 2 in cases of childhood acute lymphoblastic leukemia implicates immunosuppression in etiology. Haematologica. 2019. |
R836159 (2019) |
|
Relevant Websites:
Love in the Time of Toxicants Exit
(Rosa and Carlos Plan a Baby Exit
The Center for Integrative Research on Childhood Leukemia and the Environment Exit
Progress and Final Reports:
Original Abstract Subprojects under this Center: (EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
R836159C001 In Utero Chemical Exposures, Immune Status, and Childhood Leukemia
R836159C002 Identifying In Utero Exposures that are Risk Factors for Childhood Leukemia
R836159C003 Prenatal Exposures, Constitutive Genetics, DNA Methylation & Childhood Leukemia
The perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.