Grantee Research Project Results
2016 Progress Report: Liver MAPs
EPA Grant Number: R835737C001Subproject: this is subproject number 001 , established and managed by the Center Director under grant R835737
(EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
Center: The Morgridge Institute for Research
Center Director: Thomson, James
Title: Liver MAPs
Investigators: Thomson, James
Institution: The Morgridge Institute for Research
Current Institution: The Morgridge Institute for Research , University of Wisconsin - Madison
EPA Project Officer: Aja, Hayley
Project Period: December 1, 2014 through November 30, 2018 (Extended to November 30, 2019)
Project Period Covered by this Report: December 1, 2015 through November 30,2016
RFA: Organotypic Culture Models for Predictive Toxicology Center (2013) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
The objectives of the project are:
OBJECTIVE 1. Identify regulatory networks that control liver maturation.
OBJECTIVE 2. Screen for factors that promote human ES/iPS cell-derived hepatocyte maturation.
OBJECTIVE 3. Optimize 3D organotypic cultures of human ES/iPS cell-derived hepatocytes with mature metabolic function.
Progress Summary:
OBJECTIVE 1. Identify regulatory networks that control liver maturation. We are performing detailed time series RNA sequencing (RNA-seq) during mouse liver development to select time points during hepatocyte maturation for use in ChIP-seq against six histone modifications shown to be sufficient to identify promoters and enhancers, as well for DNase-seq to identify transcription factors binding to regulatory elements. We have completed sample collection of all time points, daily from embryonic day 17.5 to postnatal day 42 and then every 5 days from day 45 to day 60. Female mice were excluded when possible due to variation of the estrus cycle on lever physiology. All samples have been sequenced and data are being analyzed in collaboration with Dr. Sushmita Roy of our Institute to identify time points of interest as mentioned above. Due to the technical weaknesses of DNase-seq and high cell numbers required, we have focused on ATAC-seq, which is based on DNA fragmentation using a transposase instead of DNase I. The protocol is much shorter, robust, and requires only 5x104 input cells compared to 2x107 needed for DNase-seq. We currently are optimizing the protocol to use with hepatocytes. A key step is to obtain a ladder of ~200 bp bands of the fragmented and PCR multiplied DNA corresponding to nucleosomes. We have successfully optimized this step with nuclei isolated using previously reported protocol (Figure 1). We shall next perform a full sequencing and bioinformatic analysis to confirm that open chromosome elements are indeed enriched.
OBJECTIVE 2. Screen for factors that promote human ES/iPS cell-derived hepatocyte maturation. We have taken a three-pronged approach to screen for small molecules to promote maturation.
1) We previously reported generation of a dual reporter human ES cell clone that expresses tdTomato fluorescent-protein driven by the fetal hepatic gene alpha-fetoprotein (AFP) locus along with green fluorescent-protein driven by the mature hepatic gene albumin (ALB) locus to screen small molecule library to identify candidates that mature ES-derived hepatocytes. As a first and crucial step, we have developed a defined hepatocyte differentiation protocol from ES cells, which eliminates unknown and variable factors that may interfere with our screen. Cells differentiated using this protocol are morphologically uniform with a high percentage expressing AFP. There is low ALB expression as expected in immature cells (Figure 2).
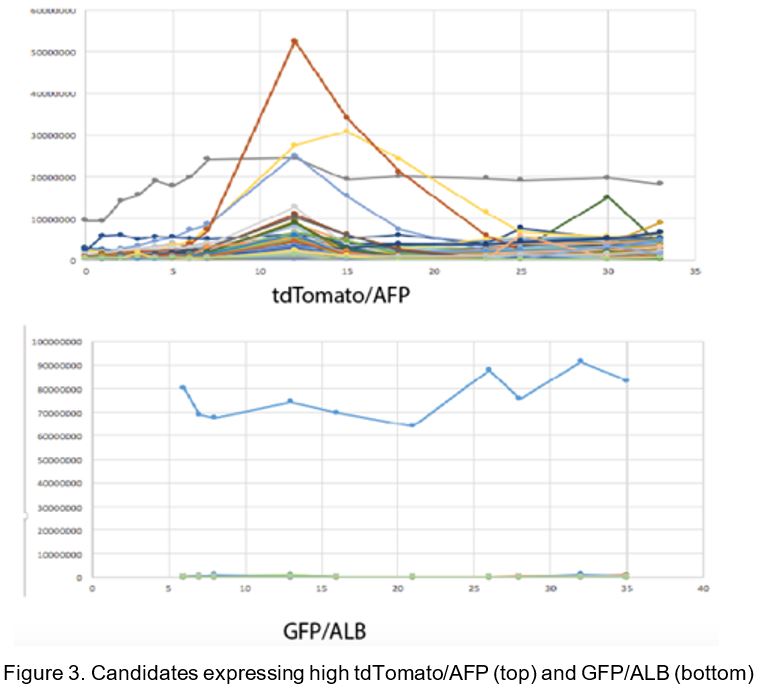
Using these cells we screened a library of 130 small molecules of known function that interact with key cellular pathways, designed in house on a customized robotic workstation (Tecan). We quantified GFP and tdTomato in the cells using an automated imaging system (Biostation CT, Nikon) and processed images with CL-Quant software (DRVision Technologies). We identified five compounds (BIO, BIO1211, ID8, INCB018424 and CP-690550) to increase AFP and 1 compound (Stauprimide) to increase ALB (Figure 3). We are in the process of following up on these candidates by RNA-seq confirmation of a larger cohort of fetal and adult hepatic genes. Our next goal is to screen a larger library of small molecules.
2) With the help of the independent contract research organization InGenious Targeting Laboratory, we have generated a targeted double knock-in mouse ES cell line with the P. pyralis luciferase gene regulated by the AFP locus and the R. reniformis luciferase gene regulated by the ALB locus. We shall use this dual reporter line to develop a defined hepatocyte differentiation protocol that we will use to screen for molecules to increase hepatic maturity using our robotic workstation and automated imaging set-up.
3) Working with InGenious Targeting Laboratory, we have generated chimeric mice using the double knock-in ES cell line described above. We received the founder mice and have established a colony. We shall next isolate hepatocytes from adult livers and use our robotic screen to identify small molecules that will help retain the maturity of adult hepatocytes in culture, where they de-differentiate and lose adult phenotype rapidly.
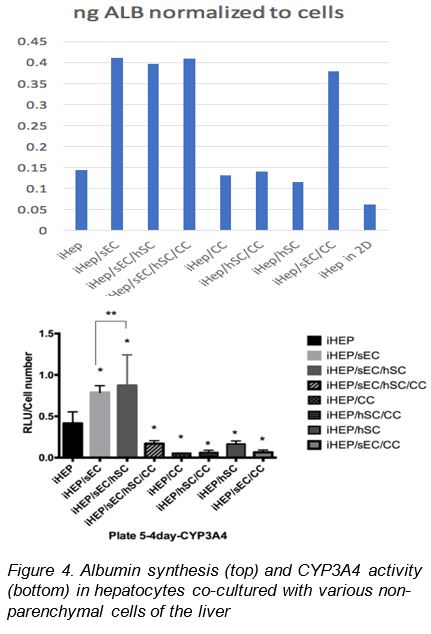
OBJECTIVE 3. Optimize 3D organotypic cultures of human ES/iPS cell-derived hepatocytes with mature metabolic function. We previously reported suspension culture of human iPS cell-derived hepatocytes (iHeps, CDI) in 3D aggregates to significantly improve metabolic activities. We have performed co-culture of iHeps with various non-parenchymal liver cells, namely, sinusoidal endothelial cells (sEC), hepatic stellate cells (hSC) and cholangiocytes (CC) in 3D to explore further improvements in metabolic functions. Albumin synthesis (as measured by secreted albumin in the media by ELISA) by iHep is significantly increased by co-culture with sEC, with or without CC and hSC. CC and hSC alone did not increase albumin synthesis (Figure 4, top panel). The activity of CYP3A4, a major metabolic enzyme (measured by Promega CYP450 Glo assay) also showed a significant increase upon co-culture with sEC. Co-culture with CC or hSC alone did not increase CYP3A4 activity (Figure 4, bottom panel). Thus, we have identified sEC to increase the maturity of iHeps in 3D aggregate co-cultures. We shall next confirm the metabolic competence of iHep/sEC 3D aggregates by studying xenobiotic metabolism by all major phase I and II enzymes.
Journal Articles:
No journal articles submitted with this report: View all 8 publications for this subprojectProgress and Final Reports:
Original AbstractMain Center Abstract and Reports:
R835737 The Morgridge Institute for Research Subprojects under this Center: (EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
R835737C001 Liver MAPs
R835737C002 Brain MAPs
R835737C003 Cancer MAPs: A 3D Organotypic Microfluidic Culture System to
Identify Chemicals that Impact Progression and Development of Breast Cancer
R835737C004 Vascular MAPs: Vascular and Neurovascular Tissue Models
R835737C005 Pathway Analysis Core
The perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
Project Research Results
4 journal articles for this subproject
Main Center: R835737
215 publications for this center
81 journal articles for this center