Grantee Research Project Results
2015 Progress Report: Epigenetics Project
EPA Grant Number: R834513C003Subproject: this is subproject number 003 , established and managed by the Center Director under grant R834513
(EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
Center: Center for Integrative Research on Childhood Leukemia and the Environment - 2015
Center Director: Metayer, Catherine
Title: Epigenetics Project
Investigators: Eskenazi, Brenda , Holland, Nina T. , Hubbard, Alan , Bradman, Asa , Harley, Kim , Barcellos, Lisa , Lustig, Robert
Current Investigators: Eskenazi, Brenda
Institution: University of California - Berkeley , University of California - San Francisco
Current Institution: University of California - Berkeley
EPA Project Officer: Hahn, Intaek
Project Period: August 1, 2009 through July 31, 2014 (Extended to July 31, 2017)
Project Period Covered by this Report: June 1, 2014 through May 31,2015
RFA: Children's Environmental Health and Disease Prevention Research Centers (with NIEHS) (2009) RFA Text | Recipients Lists
Research Category: Children's Health , Human Health
Objective:
In Project C, we are investigating molecular mechanisms with a main focus on epigenetic effects associated with prenatal and childhood exposure to persistent organic pollutants (DDT and PBDEs). We also are characterizing the differences in multiple epigenetic marks by host factors like age and sex in children from birth to 12 years of age and assessing relationships of epigenetic modifications in children with hormone levels and pubertal onset.
Progress Summary:
1. To analyze global DNA methylation in newborn children by three different assays.
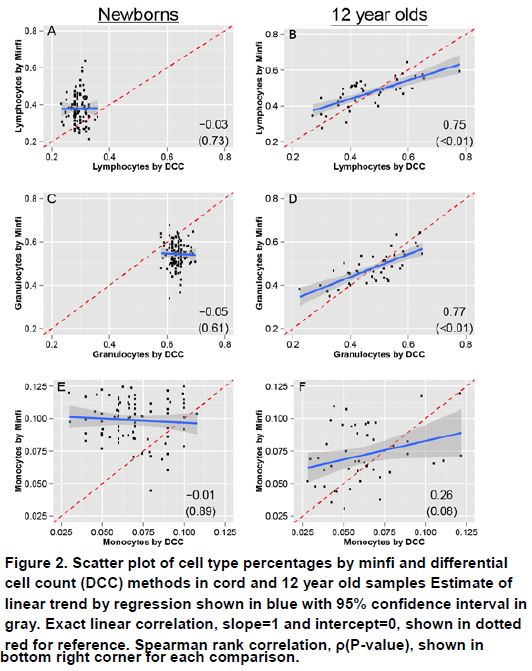
Previously, we reported on a novel method of normalization of genome-wide site-specific methylation data obtained by 450K BeadChip arrays for birth cohort and other longitudinal population studies (Yousefi et al 2013). Another important methodological question we have addressed using the 450K data from the CHAMACOS cohort is the potential confounding in methylation studies by blood composition since methylation levels can differ among white blood cell types. One of the most commonly used methods to estimate cell composition in blood specimens assessed by 450K BeadChip is an algorithm known as the Houseman adjustment that is implemented by the Bioconductor package minfi. However, the algorithm is based on the methylation profiles of six Swedish men that may not be comparable to young children because the immune profiles in young children, especially newborns, are vastly different than those of adults. We estimated white blood cell type percentages by two methods, minfi and direct differential cell count (DCC), at two time points in childhood (birth and 12 years of age). We found that both minfi and DCC had similar trends as children aged, and neither count method differed by sex among newborns (p > 0.10)(Figure 2). However, minfi estimates did not correlate well with DCC in samples from newborns (ρ = -0.05 for granulocytes; ρ = -0.03 for lymphocytes). In older children, correlation improved substantially (ρ = 0.77 for granulocytes; ρ = 0.75 for lymphocytes), likely due to increasing similarity with minfi’s adult reference data as children aged. Our findings suggest that the minfi method may provide suitable estimates of white blood cell composition for samples from adults and older children, but may not be appropriate for EWAS involving newborns or young children. A manuscript on the estimation of blood cell heterogeneity in children for epigenome-wide studies has been provisionally accepted for publication by Environmental and Molecular Mutagenesis (Yousefi et al. 2015).
All proposed analyses of global methylation (Alu, LINE-1) were completed in 246 newborns and 246 9-year old children who had blood samples available for DNA isolations. These samples, which had been previously collected and banked in the CHAMACOS study biorepository, provided a unique opportunity to explore epigenetic effects of prenatal exposure and of sex and age from birth to near the onset of puberty. We did not observe significant correlations between the two different assays used for analysis of global methylation, most likely because they characterize complementary parts of the epigenome and thus together assure its more complete evaluation. A paper on global repeats (Alu and LINE-1) has been published (Huen et al, 2014).
2. To determine ontogenetic changes in global DNA methylation in blood of children between birth and 12 years.
To determine the longitudinal changes in repetitive element DNA methylation (Alu and LINE-1), pyrosequencing assays were performed in > 600 blood samples collected from CHAMACOS children at delivery and ages 2, 5, 7 and 9. We used generalized estimating equations (GEE) to assess differences in LINE-1 and Alu methylation by age while accounting for within subject correlations. LINE-1 global DNA methylation was found to decrease 0.3% per month increase in child age (95% CI: 0.2 – 0.6%). A similar decreasing trend was found for Alu methylation but this association was not statistically significant (p = 0.18). Although these differences are not large, previous studies have shown that even small differences in methylation can be biologically meaningful. Comparison of the data for boys and girls at birth and 9 years of age has been published (Huen et al 2014). We are also finalizing a detailed age-related comparison of epigenetic marks by the 450K assay for submission to PlosONE (Yousefi et al).
3. To investigate the relationship of in utero and 9-year-old blood concentrations of DDT/E and PBDEs with global DNA methylation.
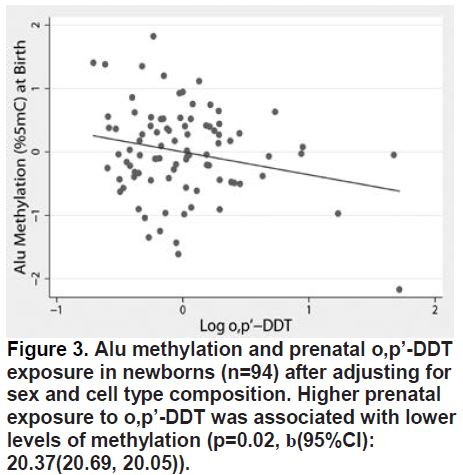
We observed a consistent trend of lower Alu and LINE-1 methylation in fetal blood with higher prenatal DDT/E exposure, particularly after adjusting for cell type composition (Figure 3).
Prenatal PBDE exposure was not significantly associated with Alu methylation. There was a trend of lower levels of LINE-1 methylation with prenatal PBDE exposures but this relationship did not reach statistical significance. Since mothers were exposed to both PBDEs and DDT/E during pregnancy, we also examined the potential association of co-exposure to these chemicals with levels of Alu and LINE-1 methylation. When we included both classes of chemicals in the same model, we found a significant interaction between sum of PBDEs and o,p’-DDT and p,p’-DDE (p = 0.12 and 0.06, respectively) on LINE-1 but not Alu methylation in fetal cord blood. Similar but non-significant interaction was seen for the sum of PBDE congeners with p,p’-DDT. These data suggest that the significant association of prenatal PBDE exposure with LINE-1 methylation in cord blood was only observed after considering exposure to both classes of compounds (PBDEs and DDT/E). Our data demonstrate that prenatal exposure to POPs may be linked to hypomethylation in fetal blood and that accounting for co-exposure to different types of chemicals and adjusting for blood cell types may increase sensitivity of epigenetic analyses for epidemiological studies. These results were recently published (Huen et al. 2014).
4. To determine whether global methylation is associated with onset of puberty and hormonal changes.
This Center grant includes the follow-up of boys only. However, we received an NIEHS grant (ES017054) to assess pubertal onset in the girls in the CHAMACOS cohort. Although we found no significant relationship between pubertal onset as determined by Tanner staging (testicular volume or pubic hair) and either LINE-1 or Alu measures in boys, we observed a non-significantly higher odds of girls entering puberty by age 9 years (Tanner stage > B1) with higher LINE-1 methylation (but not Alu methylation) in cord blood. Similarly, we also found a suggestive trend of an increased odds of having entered puberty by 9 years (Tanner stage > B1) in girls with higher Alu methylation in 9-year old blood.
We also measured concentrations of luteinizing hormone (LH), testosterone (T), and follicle stimulating hormone (FSH) in first-morning serum samples collected from the boys at 12 years that demonstrate that hormone levels in CHAM1 boys were comparable to the normal reference ranges. We found a positive albeit non-significant relationship between Alu methylation in cord blood and levels of T in boys 12 years later. In addition, we observed a significant positive relationship between LINE-1 and ln LH. We are currently finalizing a manuscript regarding the relationship of LINE-1 and Alu repetitive element methylation with onset of puberty (boys and girls) and hormonal levels (boys only) in CHAM1 children.
5. To examine site-specific methylation in relation to age, sex, exposure to DDT/E and PBDEs and puberty onset.
We observed that ~ 3% of CpG sites interrogated by the 450K BeadChip in umbilical cord blood specimens were differentially methylated between girls and boys at birth (FDR P < 0.05), after adjusting for cell composition. Of those CpGs, 3,031 were located on autosomes, and 82.9% of those were hypermethylated in girls compared to boys. Beyond individual CpGs, we found 3,604 sex-associated differentially methylated regions (DMRs) where the majority (75.8%) had higher methylation in girls. Using pathway analysis, we found that sex-associated autosomal CpGs were significantly enriched for gene ontology terms related to nervous system development and behavior. Among hits in our study, 35.9% had been previously reported as sex-associated CpG sites in other published studies. Further, for almost all replicated hits, the direction of the association with methylation was concordant with previous studies. Although there is a comprehensive meta-analysis of 76 studies using 27K site-specific methylation data, no such study has been published using the more comprehensive 450K assay. Ours is the first to look at differential methylation over regions of CpG sites and adjusting for potential confounding by blood cell composition. We recently submitted a manuscript on the associations of sex with site-specific methylation measured by 450K BeadChip assay (Yousefi et al., BMC Genomics).
We conducted a detailed analysis of the relationship of epigenetic data with the gene expression and genetic polymorphisms, that was recently published (Dave et al 2015), providing additional “proof of principal” validation to our epigenetic findings.
We previously reported associations of methylation at several CpG sites in multiple genes with in utero exposure to PBDEs. As a confirmation step, we are now performing real-time quantitative PCR (qRT-PCR) for the top 6 hits identified by the 450K methylation analysis. Total RNA was isolated from CHAMACOS newborns using mirVana miRNA Isolation Kit (Life Technologies, Waltham, MA, USA). Two different methods were used, the Applied Biosystems® (Applied Biosystems Life Technologies, Grand Island, NY, USA) TaqMan® RNA-to-Ct™ 1-Step Kit and QuantiTect SYBR Green PCR Kit (Qiagen, Valencia, CA). The expression of six genes (ITSN1, CRYZL1, HSPA2, FGF3, KCNMB4 and CREB3L1) and 2 housekeeping genes (ACTB and TBP) were measured in 94 newborns in June 2015. To complete the validation efforts, a detailed analysis of the relationship of expression with CpG-sites methylation in the same genes will be performed during the next year.
Significance:
Novel epigenetic findings on the molecular mechanisms associated with environmental exposures in newborns and children resulting from the CHAMACOS Project C were only possible because of the existing CHAMACOS birth cohort data and banked biological samples collected over the children’s lifetime and going back 15 years to the time when CHAMACOS children were in utero.
This project required a careful examination of existing methodological and analytical approaches. We conducted a direct comparison of DNA methylation in different cell types that demonstrated that the commonly used minfi procedure may not be appropriate for newborns that have a different blood cell composition than adults. We developed novel methods of data normalization of high-dimensional genome-wide methylation data that was submitted to public domain. We employed state-of the art analytical methodologies such as SuperLearner for statistical analyses. We are now applying these methods in combined analysis of the effects of maternal smoking on DNA methylation by 450K BeadChip in multiple birth cohorts, including the CHAMACOS cohort, under the Pregnancy and Childhood Consortium (PACE). The CHAMACOS cohort of Mexican-American newborns is unique and it adds important information on ethnic/racial variability in this international consortium primarily comprised of Caucasian cohorts (this paper is in preparation for Nature Genetics).
Our data show striking differences in genome-wide DNA methylation by age and sex, not limited to CpG sites located in sex chromosomes. This provides additional evidence for the need to expand research on environmental epigenetics to a wide variety of sub-populations, including to fetuses and pregnant women and minority cohorts (e.g., CHAMACOS) to explain differential risks and health effects. We first reported data in our birth cohort demonstrating that prenatal exposure to POPs such as PBDE and DDT that are still common in California and many other parts of the world may be linked to hypomethylation in fetal blood. Importantly, we showed that accounting for co-exposure to different types of chemicals and adjusting for blood cell types may increase sensitivity of epigenetic analyses for epidemiological studies. This is immediately relevant to the EPA priority science question “What chemicals and combinations of chemicals… pose the greatest risk to children’s health?”
Future Activities:
In the next year, we plan to finalize data analysis of qPCR expression experiments performed for validation of hits identified by 450K array. We have already completed all the data analyses and will submit during next year manuscripts on: (a) 450K methylation and PBDE exposure; (b) 450K methylation and age; and (c) pubertal onset and hormone levels with global methylation.
Journal Articles on this Report : 8 Displayed | Download in RIS Format
| Other subproject views: | All 77 publications | 38 publications in selected types | All 38 journal articles |
|---|---|---|---|
| Other center views: | All 697 publications | 170 publications in selected types | All 169 journal articles |
| Type | Citation | ||
|---|---|---|---|
|
|
Chadwick LH, Sawa A, Yang IV, Baccarelli A, Breakefield XO, Deng H-W, Dolinoy DC, Fallin MD, Holland NT, Houseman EA, Lomvardas S, Rao M, Satterlee JS, Tyson FL, Vijayanand P, Greally JM. New insights and updated guidelines for epigenome-wide association studies. Neuroepigenetics 2015;1:14-19. |
R834513 (2014) R834513 (2015) R834513C003 (2014) R834513C003 (2015) |
Exit Exit Exit |
|
|
Dave V, Yousefi P, Huen K, Volberg V, Holland N. Relationship between expression and methylation of obesity-related genes in children. Mutagenesis 2015;30(3):411-420. |
R834513 (2013) R834513 (2014) R834513 (2015) R834513 (Final) R834513C003 (2013) R834513C003 (2014) R834513C003 (2015) |
Exit Exit Exit |
|
|
Dave V, Street K, Francis S, Bradman A, Riley L, Eskenazi B, Holland N. Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatric Research 2016;79(6):846-854. |
R834513 (Final) R834513C002 (2016) R834513C003 (2015) |
Exit Exit Exit |
|
|
Holland N, Lizarraga D, Huen K. Recent progress in the genetics and epigenetics of paraoxonase: why it is relevant to children's environmental health. Current Opinion in Pediatrics 2015;27(2):240-247. |
R834513 (2013) R834513 (2014) R834513 (2015) R834513 (Final) R834513C003 (2013) R834513C003 (2014) R834513C003 (2015) R834513C004 (2013) |
Exit Exit Exit |
|
|
Huen K, Yousefi P, Street K, Eskenazi B, Holland N. PON1 as a model for integration of genetic, epigenetic, and expression data on candidate susceptibility genes. Environmental Epigenetics 2015;1(1):dvv003. |
R834513 (2015) R834513C003 (2015) |
Exit Exit Exit |
|
|
Yousefi P, Huen K, Schall RA, Decker A, Elboudwarej E, Quach H, Barcellos L, Holland N. Considerations for normalization of DNA methylation data by Illumina 450K BeadChip assay in population studies. Epigenetics 2013;8(11):1141-1152. |
R834513 (2012) R834513 (2013) R834513 (2014) R834513 (2015) R834513 (Final) R834513C003 (2012) R834513C003 (2013) R834513C003 (2014) R834513C003 (2015) |
Exit Exit Exit |
|
|
Yousefi P, Huen K, Quach H, Motwani G, Hubbard A, Eskenazi B, Holland N. Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environmental and Molecular Mutagenesis 2015;56(9):751-758. |
R834513 (2015) R834513C003 (2015) |
Exit Exit |
|
|
Yousefi P, Huen K, Dave V, Barcellos L, Eskenazi B, Holland N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics 2015;16:911. |
R834513C003 (2015) R834513C003 (2016) |
Exit Exit Exit |
Supplemental Keywords:
Epigenetics, methylation, DDT, DDE, PBDEs, flame retardants, manganese, maneb, puberty, Health, RFA, Scientific Discipline, INTERNATIONAL COOPERATION, Health Risk Assessment, Environmental Policy, Biology, Children's Health, Biochemistry, biological markers, harmful environmental agents, pesticide exposure, agricultural community, flame retardants, neurochemical effects, PBDE, children's vulnerablity, farmworkersRelevant Websites:
http://ehsdiv.sph.berkeley.edu/holland/index.php Exit
Progress and Final Reports:
Original AbstractMain Center Abstract and Reports:
R834513 Center for Integrative Research on Childhood Leukemia and the Environment - 2015 Subprojects under this Center: (EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
R834513C001 CHAMACOS Cohort Project: Pesticides and PBDE on Neurobehavior and Puberty
R834513C002 Project B: Exposure Project: Mn, DDT/E and PBDE Exposure to Farmworker Children
R834513C003 Epigenetics Project
R834513C004 Community Outreach and Translation Core
The perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
Project Research Results
- Final Report
- 2016 Progress Report
- 2014 Progress Report
- 2013 Progress Report
- 2012 Progress Report
- 2011 Progress Report
- 2010 Progress Report
- Original Abstract
38 journal articles for this subproject
Main Center: R834513
697 publications for this center
169 journal articles for this center