Grantee Research Project Results
2015 Progress Report: Brain MAPs
EPA Grant Number: R835737C002Subproject: this is subproject number 002 , established and managed by the Center Director under grant R835737
(EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
Center: Human Models for Analysis of Pathways (H MAPs) Center
Center Director: Murphy, William L
Title: Brain MAPs
Investigators: Ashton, Randolph S
Institution: University of Wisconsin - Madison
EPA Project Officer: Aja, Hayley
Project Period: December 1, 2014 through November 30, 2018 (Extended to November 30, 2019)
Project Period Covered by this Report: December 1, 2014 through November 30,2015
RFA: Organotypic Culture Models for Predictive Toxicology Center (2013) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
The Brain-MAPs project aims to develop a high-throughput neurotoxicity screening platform that recapitulates the diversity of regional cell phenotypes within the human central nervous system (CNS) while remaining sensitive enough to detect toxicity towards a single phenotype. As our first objective, we are creating chemically defined, standardized protocols for differentiating human pluripotent stem cells (hPSCs) into 36 tissues that span diverse CNS regions (Figs. 1A and B). This CNS model is being generated in a well plate format, and RNA-sequencing (RNA-seq) of each regional tissue will be used to develop a model-wide transcriptomic map. In our second objective, we are translating the CNS model to a microfluidic platform to simplify model derivation and enhance the tissues’ organotypic cytoarchitecture. Lastly, as our third objective, we will use CRISPR/Cas9 genome editing to create high-throughput screening assays for detecting phenotype-specific neurotoxicity using automated high content imaging.
Progress Summary:
Objective 1. To create a comprehensive CNS model, we initially proposed to differentiate hPSCs into neural tissues of 9 discrete rostrocaudal (R/C) domains, which would each be further differentiated across four dorsoventral (D/V) domains (Figs. 1A and B). The estimate of R/C domains was based on previously published neural differentiation protocols, which we are still following for deriving forebrain, midbrain, and rostral hindbrain tissues. However, our deterministic HOX patterning protocol has the potential to provide enhanced coverage of neuronal phenotype diversity in the caudal hindbrain (i.e. pons and medulla oblongata) and spinal cord. To test this hypothesis, we performed a preliminary time course differentiation/RNA-seq study across our presumptive caudal hindbrain derivation time frame with principal component analysis (PCA) and K-means clustering of the RNA-seq data by the Pathway Analysis Core (Fig. 1C). PCA revealed a dominant time-dependent axis (PC1) that can be sub-divided into 5 transcriptionally discrete domains, which is roughly consistent with the seven HOX expressing rhombomeres that form during hindbrain development. While this is scientifically novel, its inclusion in our CNS model will depend on RNA-seq associated cost. Discretization of the spinal cord’s R/C axis will remain as originally proposed since it captures the critical phenotypical diversity.
To develop D/V patterning protocols, we used hPSCs differentiated to the cervical spinal cord domain as a trial case. D/V patterning has been well studied in cervical spinal cord tissues of animal models, which provided a template for our human studies. Using our chemically defined system, we first discovered that transient Wnt signaling, not sonic hedgehog (Shh) signaling, induces NKX6.1 expression, a requisite transcription factor for patterning ventral spinal cord phenotypes (manuscript in review). Next, we conducted a bone morphogenetic protein (BMP) and Shh signaling dose response experiment to pattern neural progenitor subtypes of diverse D/V domains. Quantitative PCR of a transcription factor panel expressed by neural progenitors spanning the D/V axis was used to assess the extent of patterning. A heat map depiction of the QPCR results indicates our success in patterning diverse D/V progenitor phenotypes (Figs. 1B and D). Immunocytochemical analysis of these progenitor cultures and their terminally differentiated counterparts will be used as a final verification of our protocol’s capability to pattern neuronal tissues from diverse D/V domains.
Objective 2. Working with the Microscale Systems Core, we have demonstrated the culture of hPSC-derived forebrain neural progenitors within a microfluidic platform capable of generating a long-term gradient of 10 and 40 kDa molecules (Fig. 1E). This size range spans that of recombinant Wnt, BMP, and Shh growth factors, which we are currently using to demonstrate D/V patterning within micropatterned neural tissues. Moreover, we have developed dynamic culture substrates that enable spatiotemporal control over the neural tissues’ microscale morphology. This enables us to induce a biomimetic cytoarchitecture within the neural tissue initially, and then instruct 2-D radial expansion of the tissue’s morphology analogous to neural tube growth in vivo. We are continuing to optimize these processes to achieve de novo synthesis of tissues with cytoarchitectures akin to CNS slice cultures.
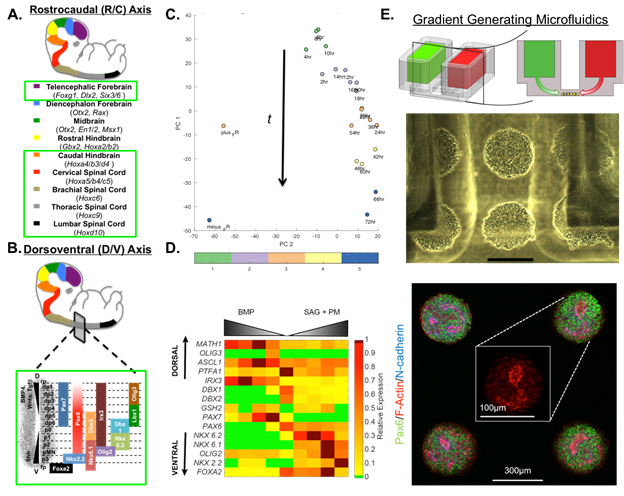
Figure 1. Brain MAPs Progress. Schematic of proposed (A) rostrocaudal and (B) dorsoventral domains that will be included in the CNS model. Standardized protocols have been developed for domains encircled in green. (C) Score plot of PCA on time course hindbrain patterning study with K-means clustering indicating 5 distinct transcriptional profiles. (D) Heat map of QPCR results from dorsoventral patterning of cervical spinal cord tissues. (E) Schematic of microfluidic platform (TOP) with cells seeded between media reservoirs (MIDDLE) containing soluble factors, 300mm scale bar. Neurally differentiating hPSCs display normal N-cadherin polarization behavior (BOTTOM) within the platform.
Future Activities:
To complete objective 1, we will standardize protocols for deriving diencephalic, midbrain, and rostral hindbrain tissues. Then, our D/V patterning protocol will be conducted at each R/C domain to complete our microwell plate CNS model, and RNA-seq will be used to develop an accompanying transcriptomic map. Additionally, we will continue to translate the well plate model to a microfluidic array of 3-D tissues with organotypic cytoarchitecture.
Journal Articles on this Report : 1 Displayed | Download in RIS Format
| Other subproject views: | All 10 publications | 4 publications in selected types | All 4 journal articles |
|---|---|---|---|
| Other center views: | All 215 publications | 82 publications in selected types | All 81 journal articles |
| Type | Citation | ||
|---|---|---|---|
|
|
Knight GT, Sha J, Ashton RS. Micropatterned, clickable culture substrates enable in situ spatiotemporal control of human PSC-derived neural tissue morphology. Chemical Communications 2015;51(25):5238-5241. |
R835737 (2015) R835737 (2016) R835737C002 (2015) R835737C002 (2016) |
Exit |
Supplemental Keywords:
Hox genes, transcription factors, neural stem cells, Paraquat, dopaminergic neuronsProgress and Final Reports:
Original AbstractMain Center Abstract and Reports:
R835737 Human Models for Analysis of Pathways (H MAPs) Center Subprojects under this Center: (EPA does not fund or establish subprojects; EPA awards and manages the overall grant for this center).
R835737C001 Liver MAPs
R835737C002 Brain MAPs
R835737C003 Cancer MAPs: A 3D Organotypic Microfluidic Culture System to
Identify Chemicals that Impact Progression and Development of Breast Cancer
R835737C004 Vascular MAPs: Vascular and Neurovascular Tissue Models
R835737C005 Pathway Analysis Core
The perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.
Project Research Results
4 journal articles for this subproject
Main Center: R835737
215 publications for this center
81 journal articles for this center
