You are here:
THE ISOLATION AND CHARACTERIZATION OF NATURALLY-OCCURRING AMOEBA-RESISTANT BACTERIA FROM WATER SAMPLES
Impact/Purpose:
Bacteria that are able to replicate and survive within amoeba hosts have been termed “amoeba-resistant bacteria” (ARB). Some of the bacteria are lytic for their amoebal hosts. These bacteria include members of the genus Legionella as well as other Legionella-like amoebal pathogens (LLAPs). Serological evidence suggests that these LLAPs may be a significant cause of respiratory disease and, because many do not grow on conventional laboratory media, they may be overlooked.
The objectives of this study are designed to address the protection of human health through clean and safe water and healthy communities. The objectives are to continue the biological clean up of previously collected water samples containing infected amoebae, continue the phylogenetic and phenotypic characterization of the bacteria, and isolate additional ARB from both environmental and human-constructed water sources.
Description:
A. Isolation of ARB from naturally-occurring amoebae and identification based on 16S rDNA sequencing.
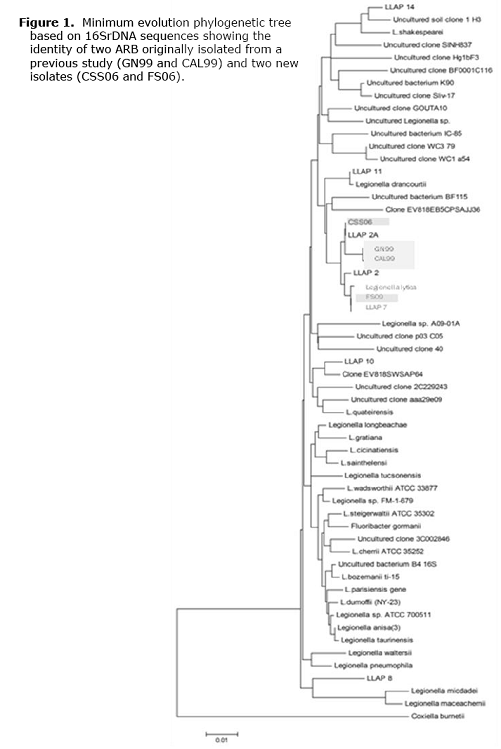
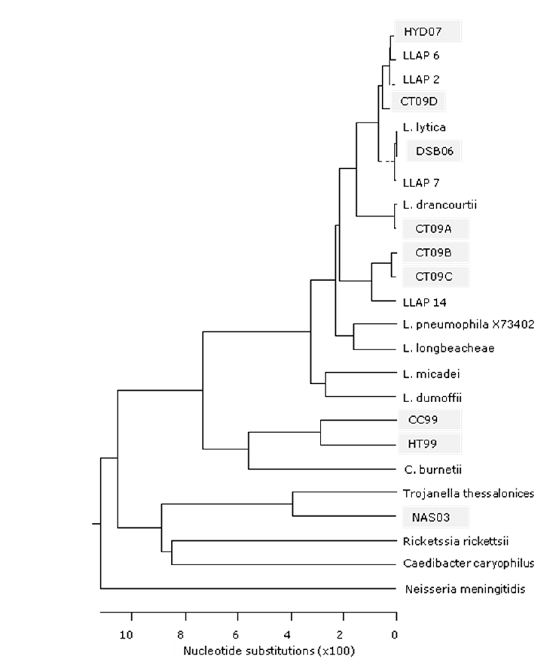
Our previous studies have resulted in the identification of several novel bacteria pathogenic for amoebae (Berk, et al. Environmental Science & Technology 2006;40:7440–7444). Our detection of the ARB differs from others in that no laboratory-grown strains of amoebae are added to enrich for the bacteria until infections in the native protozoa are observed. Because many of the infecting bacteria were unculturable on conventional laboratory media and were part of mixed infections with several other bacteria, the isolation of these organisms in axenic co-culture with laboratory strains of amoebae was challenging. From these studies, 17 ARB were successfully passaged into co-culture with Acanthamoeba polyphaga, with all but two of these organisms being nonculturable on conventional laboratory media. Eight of the ARB are in axenic co-culture with A. polyphaga without contaminating microorganisms. Sequencing of 16S rDNA has been completed on ten of the ARB (Table 1). Additionally, sequencing of four ARB from a previous study was completed and included on phylogenetic trees (Figures 1-2).
A striking result from this study was the frequency with which organisms closely related to either Legionella lytica or Legionella rowbothamii were isolated. Of the ten isolates from the present study and the four organisms characterized from a previous study, 50% were placed within this species. This is even more interesting considering that L. pneumophila was only isolated once in this study even though cooling towers were the predominant sample This suggests that L. pneumophila may not be the most frequently isolated ARB. None of these organisms were culturable on laboratory media which might explain the more predominant isolation of L. pneumophila reported by others. It is interesting that two bacteria with identity to L. shakespearei were isolated. LLAP-14, which was originally isolated in middle Tennessee, is also closely related to L. shakespearei. This may represent an ARB that is regionally predominant.
Table. 1. Characteristics of ARB in axenic co-culture with Acanthamoeba polyphaga
1 Denotes an organism isolated from a previous studied but characterized as part of current study
2 Purity of bacterium in co-culture with A. polyphaga based on absence of contaminating bacterial DNA sequences or visual eukaryotic microorganisms.
3 Identity based on closest GenBank match to the 16S rDNA sequence and the mip gene sequence for GN99, CAL99, CT09A, CT09B, CT09C, DSB06, HDY07, AND CSSO6. Identity of Legionella spp. formerly designated as LLAPs is based on Adeleke, et al. Emerging Infectious Diseases 1996;2:225-230 and Adeleke, et al. International Journal of Systemic and Evolutionary Microbiology 2001;51:1151-1160.
4 16S rDNA sequence shows less than 94% identity to either Legionella spp. or Coxiella burnetii.
5 16S rDNA sequence shows only 97% identity to Trojanella thessalonices.
6 Not determined.
7 Long-term survival of the bacteria based on their ability to infect Acanthamoeba polyphaga. ARB were co-cultured with A. polyphaga until the amoebae lysed. The lysate was sampled periodically to determine how long the bacteria remained infectious to amoebae. For survival of dessication, 50 μl samples of the lysates were placed onto empty sterile petri dishes and allowed to dry in a desiccation chamber at 5-9% relative humidity. Periodically over several months, dried lysate samples were rehydrated with sterile distilled water and fresh washed amoebae were added to the rehydrated bacteria to test for infectivity. L. pneumophila AA100 survived for 43 days in the aqueous state and for 0 days in the dessicated state.
Figure 2. Maximum parsimony phylogenetic tree based on selected 16SrDNA sequences from cultured bacteria showing the identity of three ARB originally isolated from a previous study (HT99, CC99, and NAS03) and six new isolates (DSB06, HYD07, CT09A, CT09B, CT09C, and CT09D).
B. Use of 16S rRNA gene sequences to generate probes for the distribution of the bacteria in natural and human-constructed environments.
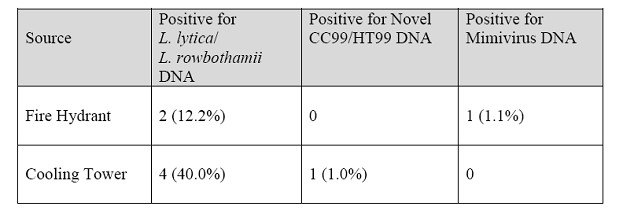
Because isolates closely related to L. lytica were isolated with the highest frequency, PCR primers unique to the 16S rDNA of this group were designed. To begin to assess the distribution of the novel species, represented by CC99 and HT99, specific primers were designed for the 16S rDNA of these organisms as well. The same samples were also screened by PCR using primers for another newly described organism, mimivirus. For these studies, DNA was extracted from water collected from 10 different cooling towers and 85 hydrant samples. All water samples were supplied with municipal water. Of the 10 cooling towers, 4 were positive for L. lytica DNA and one for DNA of the Coxiella-like organisms, HT99 and CC99. Two of 9 fire hydrant samples were positive for the L. lytica clade and one sample was positive for mimivirus DNA as confirmed by nested PCR (Table 2). This may represent the first reported detection of mimivirus in the United States.
DNA from organisms belonging to the clade containing L. lytica and L. rowbothamii was most predominant in cooling towers. This may indicate that conditions in cooling towers select for these organisms. The new species, represented by CC99 and HT99, was detected in only one of the 19 samples screened. Although a small number of samples were tested, this organism may be less populous in aqueous environments compared to other ARB. This could explain the scarcity of DNA sequences for this group in GenBank (12 sequences with >95% identity) all of which are from uncultured samples.
Table 2. Water samples positive for DNA by PCR for 16S rDNA.*
*PCR was performed using primers that could not distinguish between bacteria closely related to L. lytica/L. rowbothamii. Primers for mimivirus are from La Scola, et al. Emerging Infectious Diseases 2005;11:449-452.
C. Stability of the novel ARB outside of host cells and potential for human disease.
The majority of ARB isolates from this study are related to Legionella pneumophila, but are not culturable outside the amoeba host. Because these isolates do not replicate without a eukaryotic host, we wanted to compare their survival outside of a host with the survival of L. pneumophila.
Of the ten isolates tested (Table 1 above), only 1 had a survival pattern similar to that of L. pneumophila, which remained infectious to amoebae in a hydrated state for 43 days, but did not survive 1 day in the desiccated state. Eight of the ten isolates survived in the aqueous state longer than L. pneumophila (in the range of 50 to 130 days), and one survived up to 35 days to date. All but one of the isolates survived much longer than L. pneumophila in the desiccated state. Most were still infectious in the range of 38 to 91 days, and 2 isolates were still infectious after 15 months. The majority of the isolates were 99-100% similar to L. rowbothamii.
All of the bacteria characterized in this study reproduced in amoebae (Figure 3) and completely destroyed all amoeba cells in a co-culture within five days. Isolates CC99, HT99, NAS03, DSB06, CT09A, CT09B, and CT09C were all tested for the ability to replicate in human cells. Of all the isolates, only CC99 replicated in human cells (U937 and HeLa; Figure 4) as assessed by staining and qPCR measurement of increases in DNA. Two of the isolates, HT99 and CT09A (L. drancourtii), showed some cytopathogenicity for U937 cells but no replicating intracellular bacteria were detected.
Figure 3. Giemsa stains of selected isolates in Acanthamoeba polyphaga at 24-48 h post infection. Isolate HT99 is in the nucleus
Figure 4. DIC (left) and Giemsa stain of CC99 infecting the nuclei of human U937 macrophage-like cells and HeLa epithelial cells. Cells are completely lysed by 6 days post infection.
URLs/Downloads:
2009 Progress Report2008 Progress Report
Final Progress Report
2007 Progress Report